CUTANA CUT&RUN Library Prep Kit, Primer Set 1
High Fidelity Library Generation with Best-inclass NEBNext® Reagents
Product Description
The CUTANA™ Library Prep Kit is the first library prep kit specifically developed for CUT&RUN assays.
- Protocol is uniquely optimized for CUT&RUN
- Workflow is robust for the limited inputs generated by CUT&RUN
- Kit contains everything you need for CUT&RUN library
- Can be easily paired with the CUTANA™ CUT&RUN or CUT&RUN Protocol
- Effective for preparing CUT&RUN DNA libraries for both histone PTMs and chromatin-associated proteins
- CUTANA CUT&RUN Library Prep Kit with Multiplexing Primer Set 2
The CUTANA™ CUT&RUN Library Prep Kit offers high fidelity library generation for Illumina® sequencing by harnessing the power of New England Biolabs® best-inclass NEBNext® reagents. The kit offers a streamlined protocol specifically optimized for high sensitivity CUT&RUN applications, including those with low cell inputs.
Included are all necessary reagents to perform end repair, adaptor ligation, combinatorial dual indexing for multiplexing up to 48 samples, and DNA cleanup with SPRIselect reagent from Beckman Coulter, Inc. (see Additional Information). If additional multiplexing is desired, Primer Sets 1 and 2 can be combined for sequencing of up to 96 samples in a single lane. Pairing this kit with the EpiCypher CUT&RUN Kit affords users a cells-to-sequencing solution for chromatin mapping experiments with all the necessary controls and validated reagents to ensure confidence in obtaining high quality data. To place bulk orders, please contact us.
Validation Data
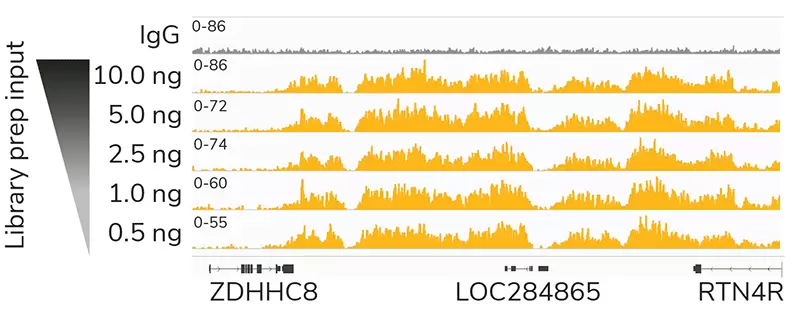
Figure 1: High quality sequencing libraries from as little as 0.5 ng of input
Gene browser shots generated using the Integrative Genomics Viewer (IGV, Broad Institute) show a representative 137 kb window for H3K27me3 antibody (ABclonal A16199) tested in CUT&RUN using 500k K562 cells. Decreasing inputs of CUT&RUN enriched DNA were used in the library prep protocol to simulate low abundance targets or low cell input experiments. Data are largely indistinguishable across inputs, demonstrating robust preparation of Illumina® NGS-ready libraries.
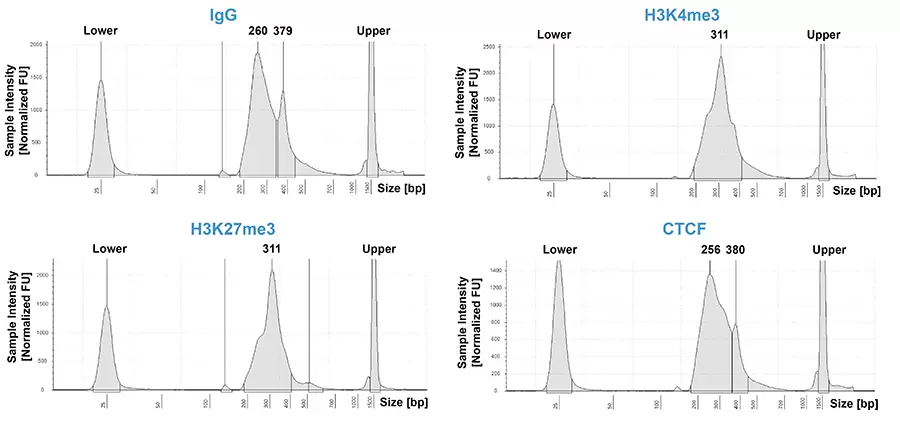
Figure 2: CUT&RUN DNA Fragment Size Distribution Analysis.
CUT&RUN was performed using the CUTANA ChIC/CUT&RUN Kit starting with 500k K562 cells. Five nanograms of CUT&RUN output DNA from reactions utilizing IgG , H3K4me3, H3K27me3 (ABclonal A16199), and CTCF antibodies were prepared for paired-end Illumina® sequencing with the CUTANA CUT&RUN Library Prep Kit. Library DNA was analyzed by Agilent TapeStation®, which confirmed that mononucleosomes were predominantly enriched in CUT&RUN (~300 bp peaks represent 150 bp nucleosomes + sequencing adapters). Peaks at ~380 bp correspond to the SNAP-CUTANA™ K-MetStat Panel of spike-in controls.
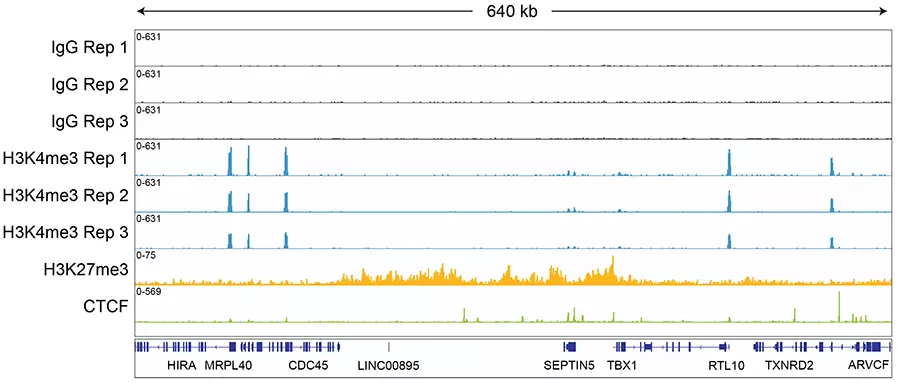
Figure 3: Representative gene browser tracks.
CUT&RUN sequencing libraries described in Figure 2 were sequenced on an Illumina® NextSeq 2000 with a P3 cartridge (paired-end 2x50 cycle). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and blacklist regions. A representative 640 kb window at the SEPTIN5 gene is shown for three replicates ("Rep") of IgG and H3K4me3 antibodies, as well as individual tracks for H3K27me3 and the transcription factor CTCF, demonstrating the robustness and reproducibility of the workflow with a variety of targets. Sequencing libraries prepared with the CUTANA CUT&RUN Library Prep kit produced the expected genomic distribution for each target. Sample sequencing depth was as follows (millions of reads): IgG Rep 1 (20.1), IgG Rep 2 (16.6), IgG Rep 3 (16.1), H3K4me3 Rep 1 (7.3), H3K4me3 Rep 2 (17.2), H3K4me3 Rep 3 (13.8), H3K27me3 (13.4), CTCF (16.2). Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute).
Related Products
CUTANA CUT&RUN Library Prep Kit with Multiplexing Primer Set 2
CUTANA pAG-MNase for ChIC/CUT&RUN Workflows
CUTANA ChIC/CUT&RUN Kit
Storage and Stability
DO NOT FREEZE ENTIRE KIT. Remove SPRIselect reagent and 0.1X TE buffer, which should be stored at RT. The remainder of the components should be stored at -20°C. Kit components stable for 6 months from date of receipt.
Additional Information
Beckman Coulter, the stylized logo, and SPRIselect are trademarks or registered trademarks of Beckman Coulter, Inc. in the United States and other countries.
Product Citations
- Andrea Berardi, Charlotte Leonie Kaestner, Michela Ghitti, Giacomo Quilici, Paolo Cocomazzi, Jianping Li, Federico Ballabio, Chiara Zucchelli, Stefan Knapp, Jonathan D Licht, Giovanna Musco (2025) The C-terminal PHD V C5HCH tandem domain of NSD2 is a combinatorial reader of unmodified H3K4 and tri-methylated H3K27 that regulates transcription of cell adhesion genes in multiple myeloma Nucleic Acids Research
- Jiaying Chen, Na Liu, Hongying Qi, Nils Neuenkirchen, Yuedong Huang, Haifan Lin (2025) Piwi regulates the usage of alternative transcription start sites in the Drosophila ovary Nucleic Acids Research
- Starble Rebecca M., Sun Eric G., Gbyli Rana, Radda Jonathan, Lu Jiuwei, Jensen Tyler B., Sun Ning, Khudaverdyan Nelli, Hu Bomiao, Melnick Mary Ann, Zhao Shuai, Roper Nitin, Wang Gang Greg, Song Jikui, Politi Katerina, Wang Siyuan, Xiao Andrew Z. (2025) Epigenetic priming promotes acquisition of tyrosine kinase inhibitor resistance and oncogene amplification in human lung cancer bioRxiv
- Xie Yipeng, Lim Jun Yi Stanley, Liu Wenyue, Gilbreath Collin, Kim Yoon Jung, Wu Sihan (2025) Transcription Machinery Anchors ecDNAs to Mitotic Chromosomes for Segregation bioRxiv
- Alison K Francois, Ali Rohani, Matt Loftus, Sara Dochnal, Joel Hrit, Steven McFarlane, Abigail Whitford, Anna Lewis, Patryk Krakowiak, Chris Boutell, Scott B Rothbart, David Kashatus, Anna R Cliffe (2024) Single-genome analysis reveals a heterogeneous association of the herpes simplex virus genome with H3K27me2 and the reader PHF20L1 following infection of human fibroblasts. mBio
- Catalog Number
14-1001-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

