Rabbit IgG Antibody, CUTANA™ CUT&RUN Negative Control
CUTANA™ CUT&RUN kits, reagents, and assay services map histone PTMs and chromatin-interacting proteins with high resolution, at a fraction of the time and cost of standard ChIP-seq experiments.
Product Description
This Rabbit IgG Antibody can be used to control for non-specific background signal in CUT&RUN and other applications. A positive control antibody is also available (Histone H3K4me3 CUT&RUN Positive Control Antibody).
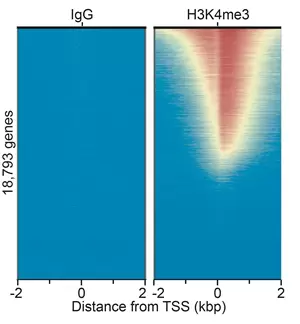
Figure 1. Heatmap for Rabbit IgG CUT&RUN negative control antibody (left) compares to Histone H3K4me3 CUT&RUN positive control antibody (right). Signal from 2.2 and 3.9 million paired-end reads, repectively, are aligned to the transcription start site (TSS, +/- 2kb) of 18,793 genes. High and low signal are ranked by intensity (top to bottom) and reflected by red and blue colors, respectively.
CUT&RUN data was generated using the EpiCypher CUTANA™ CUT&RUN protocol and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target protein.
Product Citations
- Lucie Darmusey, Anna J Bagley, Thai T Nguyen, Hanqian L Carlson, Hunter Blaylock, Shawn B Shrestha, Amara Pang, Samantha Tauchmann, Sarah C Taylor, Amy C Foley, Katia E Niño, Eric M Pietras, Theodore P Braun, Julia E Maxson (2025) Dual ASXL1 and CSF3R mutations drive myeloid biased stem cell expansion and enhance neutrophil differentiation. Blood advances
- Starble Rebecca M., Sun Eric G., Gbyli Rana, Radda Jonathan, Lu Jiuwei, Jensen Tyler B., Sun Ning, Khudaverdyan Nelli, Hu Bomiao, Melnick Mary Ann, Zhao Shuai, Roper Nitin, Wang Gang Greg, Song Jikui, Politi Katerina, Wang Siyuan, Xiao Andrew Z. (2025) Epigenetic priming promotes acquisition of tyrosine kinase inhibitor resistance and oncogene amplification in human lung cancer bioRxiv
- Turano Paolo S., Akbulut Elizabeth, Dewald Hannah K., Vasilopoulos Themistoklis, Fitzgerald-Bocarsly Patricia, Herbig Utz, MartÃnez-Zamudio Ricardo Iván (2025) Epigenetic mechanisms regulating CD8+ T cell senescence in aging humans bioRxiv
- Umeda Masayuki, Hiltenbrand Ryan, Michmerhuizen Nicole L., Barajas Juan M., Thomas Melvin E., Arthur Bright, Walsh Michael P, Song Guangchun, Ma Jing, Westover Tamara, Kumar Amit, Pölönen Petri, Mecucci Cristina, Giacomo Danika Di, Locatelli Franco, Masetti Riccardo, Bertuccio Salvatore N., Pigazzi Martina, Pruett-Miller Shondra M., Pounds Stanley, Rubnitz Jeffrey, Inaba Hiroto, Papadopoulos Kyriakos P., Wick Michael J., Iacobucci Ilaria, Mullighan Charles G., Klco Jeffery M. (2025) Fusion oncoproteins and cooperating mutations define disease phenotypes in NUP98 -rearranged leukemia medRxiv
- Xie Yipeng, Lim Jun Yi Stanley, Liu Wenyue, Gilbreath Collin, Kim Yoon Jung, Wu Sihan (2025) Transcription Machinery Anchors ecDNAs to Mitotic Chromosomes for Segregation bioRxiv
- Catalog Number
13-0042-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

