Histone H3K4me3 Antibody: SNAP-ChIP Certified, CUTANA CUT&RUN Compatible
CUTANA Compatible Antibodies meet the criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping.
Product Description
This antibody meets EpiCypher's lot-specific SNAP-Certified™ criteria for specificity and efficient target enrichment in both CUT&RUN and ChIP applications. In CUT&RUN, this requires <20% cross reactivity to related histone PTMs determined using the SNAP-CUTANA™ K-MetStat Panel of spike-in controls (19-1002-EPC). High target efficiency is confirmed by consistent genomic enrichment at 500k and 50k starting cells. In ChIP, this requires <20% cross-reactivity to related histone PTMs and >5% of target input recovered as determined using the SNAP-ChIP® K-MetStat Panel of spike-in controls (19-1001-EPC)
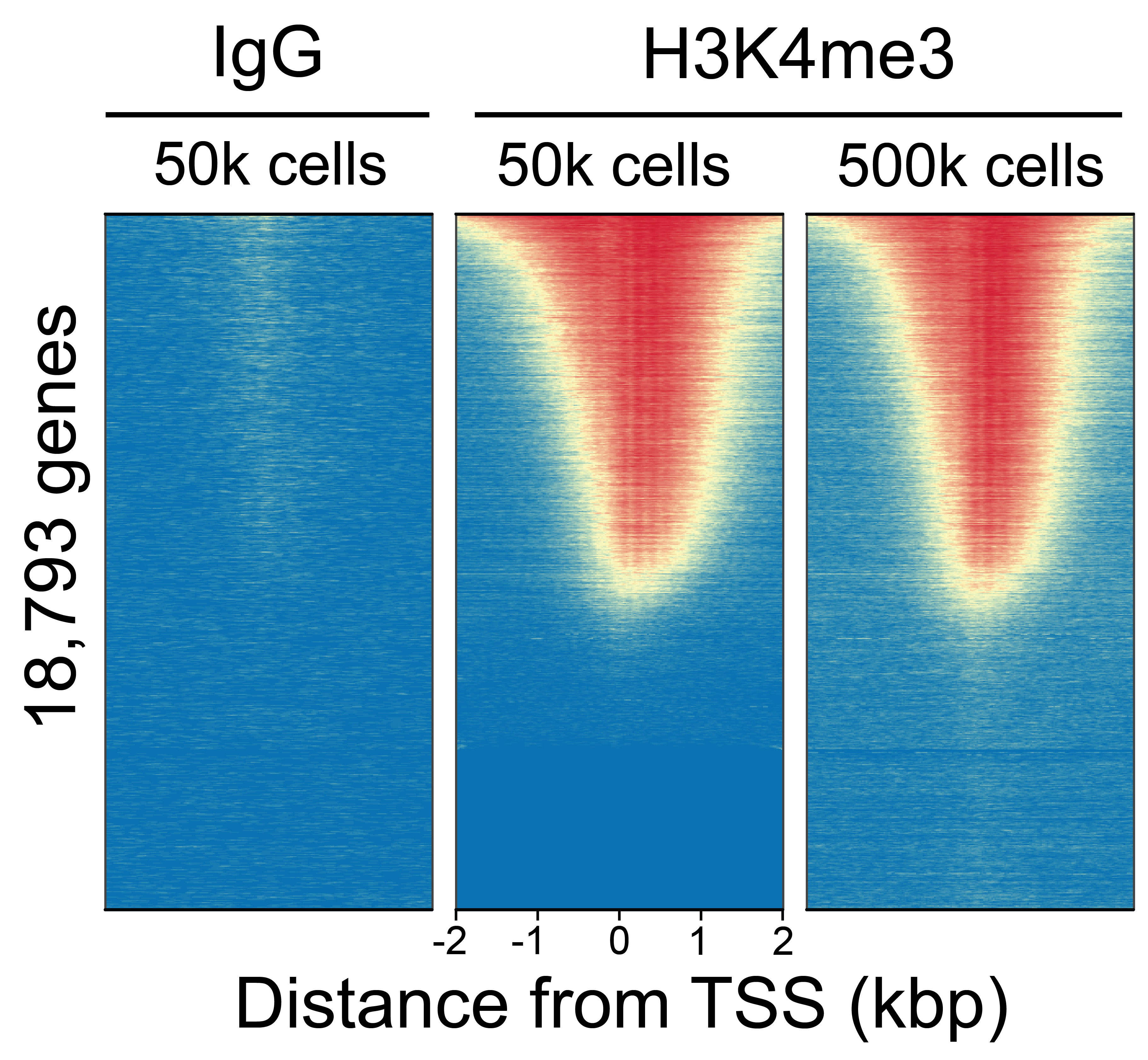
Figure 1: CUT&RUN genome wide enrichment
CUT&RUN was performed as above (Figure 1). Sequence reads were aligned to 18,793 annotated transcription start sites (TSSs, ± 2 kbp). Signal enrichment was sorted from highest to lowest (top to bottom) relative to H3K4me3 - 50k sample (all gene rows aligned). High, medium, and low intensity are shown in red, yellow, and blue, respectively. H3K4me3 antibody produced the expected TSS enrichment pattern that was consistent between 500k and 50k cells, while IgG negative control antibody (13-0042-EPC) displayed minimal background.
References
Grzybowski et al (2015) Mol Cell 58:886
Shah et al (2018) Mol Cell 72:162
Product Citations
- Madhavi Murali, Abbas Saeed, Sohyoung Kim, Kyunghee Burkitt, Hui Cheng, Arfa Moshiri, Jawad Akhtar, Daniel Tsai, Marie Luff, Baktiar Karim, Vassiliki Saloura (2025) SMYD3 drives cell cycle and epithelial-mesenchymal transition pathways through dual gene transcriptional repression and activation in HPV-negative head and neck cancer Scientific Reports
- Qingfei Zheng, Benjamin H. Weekley, David A. Vinson, Shuai Zhao, Ryan M. Bastle, Robert E. Thompson, Stephanie Stransky, Aarthi Ramakrishnan, Ashley M. Cunningham, Sohini Dutta, Jennifer C. Chan, Giuseppina Di Salvo, Min Chen, Nan Zhang, Jinghua Wu, Sasha L. Fulton, Lingchun Kong, Haifeng Wang, Baichao Zhang, Lauren Vostal, Akhil Upad, Lauren Dierdorff, Li Shen, Henrik Molina, Simone Sidoli, Tom W. Muir, Haitao Li, Yael David, Ian Maze (2025) Bidirectional histone monoaminylation dynamics regulate neural rhythmicity Nature
- Umeda Masayuki, Hiltenbrand Ryan, Michmerhuizen Nicole L., Barajas Juan M., Thomas Melvin E., Arthur Bright, Walsh Michael P, Song Guangchun, Ma Jing, Westover Tamara, Kumar Amit, Pölönen Petri, Mecucci Cristina, Giacomo Danika Di, Locatelli Franco, Masetti Riccardo, Bertuccio Salvatore N., Pigazzi Martina, Pruett-Miller Shondra M., Pounds Stanley, Rubnitz Jeffrey, Inaba Hiroto, Papadopoulos Kyriakos P., Wick Michael J., Iacobucci Ilaria, Mullighan Charles G., Klco Jeffery M. (2025) Fusion oncoproteins and cooperating mutations define disease phenotypes in NUP98 -rearranged leukemia medRxiv
- André Bortolini Silveira, Alexandre Houy, Olivier Ganier, Begüm Özemek, Sandra Vanhuele, Anne Vincent-Salomon, Nathalie Cassoux, Pascale Mariani, Gaelle Pierron, Serge Leyvraz, Damian Rieke, Alberto Picca, Franck Bielle, Marie-Laure Yaspo, Manuel Rodrigues, Marc-Henri Stern (2024) Base-excision repair pathway shapes 5-methylcytosine deamination signatures in pan-cancer genomes. Nature communications
- Arun Padmanabhan, Yvanka de Soysa, Angelo Pelonero, Valerie Sapp, Parisha P. Shah, Qiaohong Wang, Li Li, Clara Youngna Lee, Nandhini Sadagopan, Tomohiro Nishino, Lin Ye, Rachel Yang, Ashley Karnay, Andrey Poleshko, Nikhita Bolar, Ricardo Linares-Saldana, Sanjeev S. Ranade, Michael Alexanian, Sarah U. Morton, Mohit Jain, Saptarsi M. Haldar, Deepak Srivastava, Rajan Jain (2024) A Genome-Wide CRISPR Screen Identifies BRD4 as a Regulator of Cardiomyocyte Differentiation Nature cardiovascular research
- Catalog Number
13-0041-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

