SNAP-CUTANA K-MetStat Panel
First-in-class Quantitative Controls for Epigenomic Analysis
Product Description
- A direct, quantitative readout of experimental success
- In-assay validation of antibody specificity
- Robust normalization for cross-sample comparisons
- Confidence in your results with one simple spike-in addition step
The SNAP-CUTANA K-MetStat Panel of spike-in controls for CUT&RUN and CUT&Tag offers an all-in-one solution to determine antibody specificity for histone posttranslational modifications (PTMs), monitor assay success, and normalize data for quantitative chromatin mapping. The panel contains designer nucleosomes (dNucs) representing 16 different K-methyl PTM states: mono-, di-, and trimethylation at H3K4, H3K9, H3K27, H3K36, & H4K20, as well as unmodified control (Figure 1). Each PTM is represented by two unique DNA-barcoded templates (A and B, for an internal technical replicate). Each dNuc is individually conjugated to paramagnetic beads and pooled into a single panel for convenient one-step spike-in to CUT&RUN and CUT&Tag experiments. The panel is added to samples alongside ConA-immobilized cells prior to the addition of an antibody targeting a histone lysine methylation state or IgG negative control (see Table 1). pAG-MNase-mediated release or pAG-Tn5-mediated tagmentation of genomic chromatin and the barcoded nucleosomes is dependent on the specificity of the antibody used. See also SNAP-CUTANA™ Spike-in User Guide for more detailed information.
Formulation
A mixture of 16 PTM-defined semi-synthetic nucleosomes conjugated to paramagnetic beads in 10 mM sodium cacodylate pH 7.5, 100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor Cocktail, 100 μg/mL BSA, 10 mM β-mercaptoethanol. Molarity = 3 nM each, 96 nM total nucleosome. Average MW = ~263,072.7 Da.
Storage and Stability
Store at -20°C. Lower temperatures can cause freezing and will permanently damage the magnetic beads. Stable for six months from date of receipt.
Validation Data
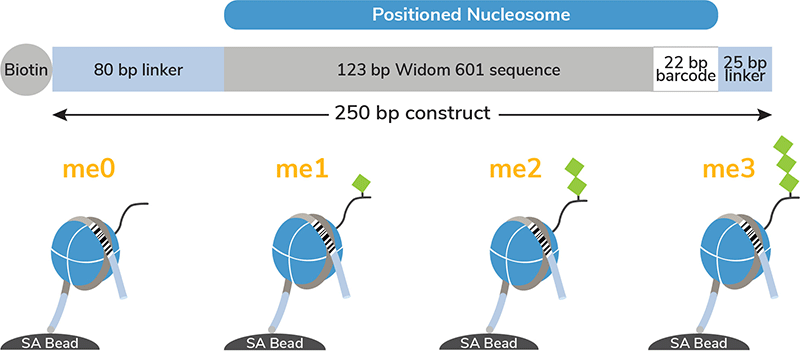
Figure 1: Schematic of SNAP-CUTANA Spike-in Controls. The K-MetStat Panel controls for 15 widely studied methyl states (see Description) and an unmodified control (me0), with each of the 16 octamers wrapped with two uniquely barcoded DNA templates (A and B). Each 250 bp DNA template contains a 123 bp 601 nucleosome positioning sequence (gray) [1], a unique 22 bp DNA-barcode (white; 32 barcodes total), and a 5’ biotin-TEG. The 5’ and 3’ linkers (blue) are compatible with cleavage by pAG-MNase (14-1048-EPC, 15-1016-EPC) during CUT&RUN as well as tagmentation by pAG-Tn5 (15-1017-EPC) during CUT&Tag. The dNucs are individually pre-conjugated to paramagnetic beads and pooled for convenient use.
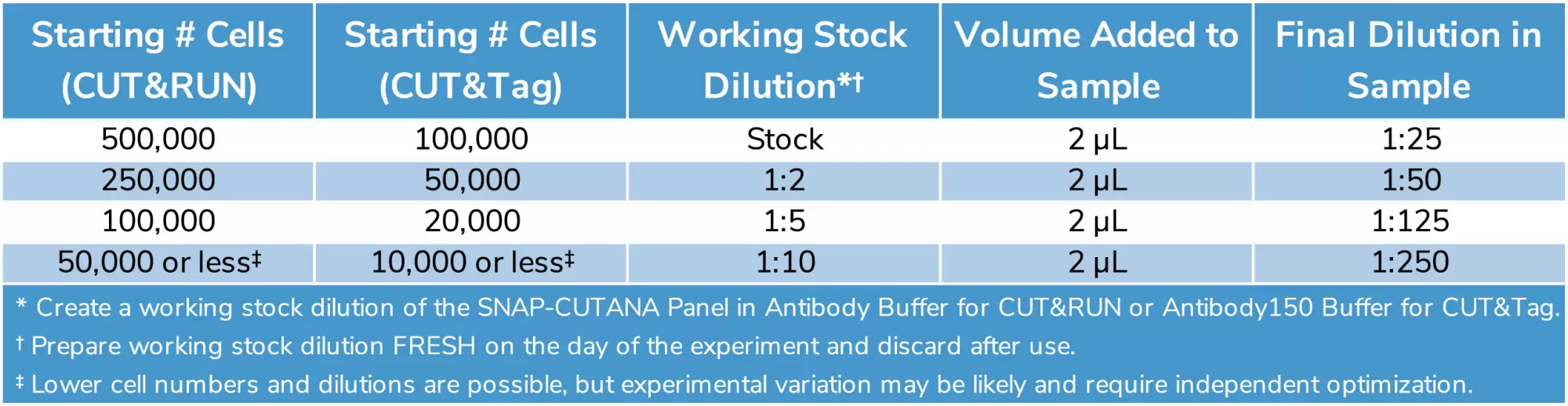
Table 1: Recommended SNAP-CUTANA Spike-In Dilution for CUT&RUN / CUT&Tag Samples of Varying Starting Cell Number
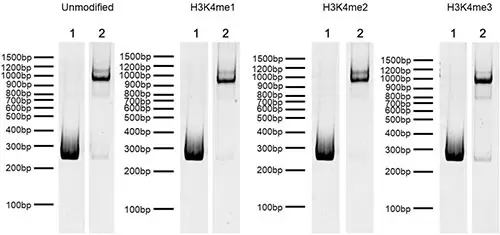
Figure 4: DNA Gel Data. Representative images for SNAP-CUTANA K-MetStat nucleosomes resolved by native PAGE and stained with ethidium bromide to confirm intact nucleosome assembly with minimal free DNA. Lane 1: Free 250 bp DNA used in nucleosome assembly (100 ng). Lane 2: Intact nucleosomes (400 ng). Comparable experiments were performed for the entire SNAP-CUTANA K-MetStat Panel.
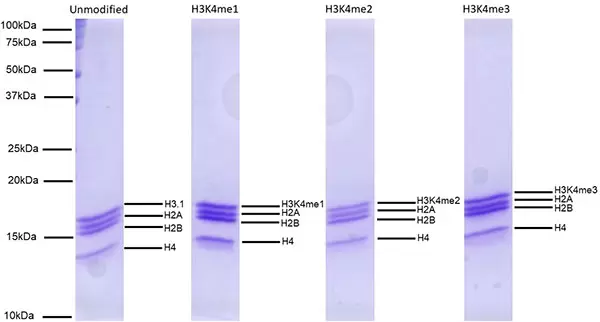
Figure 5: Protein Gel Data. Representative Coomassie stained PAGE of SNAP-CUTANA K-MetStat dNucs (1 µg each) demonstrates the purity of histones in the preparation. Sizes of molecular weight markers and positions of the core histones (H2A, H2B, H3 and H4) are indicated. Comparable experiments were performed for the entire SNAP-CUTANA K-MetStat Panel.
Related Products
The SNAP-CUTANA K-MetStat Panel of spike-in controls is suitable for CUTANA CUT&RUN Assays and CUTANA CUT&Tag Assays.
Product Citations
- Yaqiong Liu, Xianzhong Lau, Prabhakaran Munusamy, Carlos M Abascal Sherwell Sanchez, Daniel Snell, Mahesh Sangrithi (2025) Single-cell RNA-seq identifies protracted mouse germline X chromosome reactivation dynamics directed by a PRC2-dependent mechanism. Developmental cell
- Firestone Tessa M., Venters Bryan J., Novitzky Katherine, Albertorio-Sáez Liz Marie, Barnes Courtney A., Fedder-Semmes Karlie N., Hall Nathan W., Hickman Allison R., Kaderli Mark, Windham Carolina Lin, Marunde Matthew R., Maryanski Danielle N., Noll Kelsey, Shannon Leslie, Spengler Jennifer, Cowles Martis W., Sun Zu-Wen, Keogh Michael-Christopher, Johnstone Andrea L., Weinzapfel Ellen N., Sun Lu (2024) High-efficiency genomic mapping of chromatin-associated targets with CUT&RUN bioRxiv
- Giuseppe Leuzzi, Alessandro Vasciaveo, Angelo Taglialatela, Xiao Chen, Tessa M Firestone, Allison R Hickman, Wendy Mao, Tanay Thakar, Alina Vaitsiankova, Jen-Wei Huang, Raquel Cuella-Martin, Samuel B Hayward, Jordan S Kesner, Ali Ghasemzadeh, Tarun S Nambiar, Patricia Ho, Alexander Rialdi, Maxime Hebrard, Yinglu Li, Jinmei Gao, Saarang Gopinath, Oluwatobi A Adeleke, Bryan J Venters, Charles G Drake, Richard Baer, Benjamin Izar, Ernesto Guccione, Michael-Christopher Keogh, Raphael Guerois, Lu Sun, Chao Lu, Andrea Califano, Alberto Ciccia (2024) SMARCAL1 is a dual regulator of innate immune signaling and PD-L1 expression that promotes tumor immune evasion. Cell
- Haikuo Li, Dian Li, Benjamin D. Humphreys (2024) Chromatin conformation and histone modification profiling across human kidney anatomic regions Scientific Data
- Hegde Samarth, Giotti Bruno, Soong Brian Y., Halasz Laszlo, Berichel Jessica Le, Magen Assaf, Kloeckner Benoit, Mattiuz Raphaël, Park Matthew D., Marks Adam, Belabed Meriem, Hamon Pauline, Chin Theodore, Troncoso Leanna, Lee Juliana J., Ahimovic Dughan, Bale Michael, Chung Grace, D’souza Darwin, Angeliadis Krista, Dawson Travis, Kim-Schulze Seunghee, Flores Raja M., Kaufman Andrew J., Ginhoux Florent, Josefowicz Steven Z., Ma Sai, Tsankov Alexander M., Marron Thomas U., Brown Brian D., Merad Miriam (2024) Myeloid progenitor dysregulation fuels immunosuppressive macrophages in tumors bioRxiv
References
- Lowary & Widom (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning J. Mol. Biol.
- Catalog Number
19-1002-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

