CUTANA™ pAG-Tn5 for CUT&Tag
A Fusion of Proteins A and G to Transposase Tn5
Product Description
Recombinantly produced in E. coli, CUTANA pAG-Tn5 for CUT&Tag is a fusion of Proteins A and G to Transposase Tn5. This construct is useful in performing Cleavage Under Targets and Tagmentation (CUT&Tag). The active dimer of Transposase Tn5 is charged with Illumina adapters and ready to be used immediately in CUT&Tag. CUTANA pAG-Tn5 does not contain an epitope tag, which makes it compatible with tag-mediated CUT&Tag (e.g. FLAG, HA, TY1, etc.).
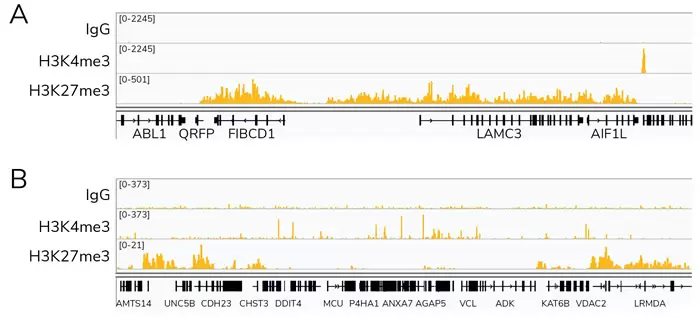
Fig. 1: CUT&Tag Data. Representative sequencing tracks obtained using CUTANA pAG-Tn5 are shown for two different loci: a 300 kb close up view of the LAMC3 gene (A) and a wide 5,608 kb window (B). CUT&Tag was performed using 100,000 K-562 nuclei with H3K4me3 antibody (EpiCypher Catalog No. 13-0041, Lot 20083002-42), H3K27me3 antibody (EpiCypher Catalog No. 13-0030, Lot 18303001) and negative control Rabbit IgG (EpiCypher Catalog No. 13-0042, Lot 20036001-52). Total paired-end read counts were 0.7, 1.1, and 0.9 million reads for H3K4me3, H3K27me3, and IgG, respectively. Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute). CUTANA pAG-Tn5 produced clear peaks with genomic distribution profiles consistent with the known biological functions of H3K4me3 and H3K27me3.
Further products from EpiCypher for CUT&Tag Assays are: CUTANA™ High Fidelity 2X PCR Master Mix, Anti-Mouse Secondary Antibody and the Anti-Rabbit Secondary Antibody.
Application Notes
Recommended use: 2.5 μL of the supplied enzyme into a 50 μL CUT&Tag reaction (20X dilution). For detailed applications and uses of this product, please see CUT&Tag protocol at: EpiCypher optimized protocols .
Adapter Sequences:
Tn5ME-A: 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3’
Tn5ME-B: 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3’
Tn5MErev: 5’-[phos]CTGTCTCTTATACACATCT-3’
NOTE: Since CUT&Tag has lower background and is compatible with fewer cells compared to ChIP-seq, it is not recommended to assess fragment size distribution using agarose gel or capillary electrophoresis (e.g. Agilent Bioanalyzer or TapeStation) prior to library preparation. This analysis is not indicative of the success of a CUT&Tag experiment, and further the amount of DNA recovered may be below the sensitivity of detection for these approaches. To gauge the success of a CUT&Tag experiment, assess DNA yield compared to positive (e.g. H3K4me3, H3K27me3) and negative (IgG) controls, determine fragment size distribution of sequence-ready libraries, and evaluate peak structure and expected genome-wide distribution in NGS data.
References
(1) Kaya-Okur et. al, Nat. Commun. 2019 (PMID : 31036827)
(2) Henikoff and Henikoff, bioRxiv. 2020 (2020.04.15.043083)
Product Citations
- Kazumi Shimaoka, Kei Hori, Satoshi Miyashita, Yukiko U Inoue, Nao K N Tabe, Asami Sakamoto, Ikuko Hasegawa, Kayo Nishitani, Kunihiko Yamashiro, Saki F Egusa, Shoji Tatsumoto, Yasuhiro Go, Manabu Abe, Kenji Sakimura, Takayoshi Inoue, Takuya Imamura, Mikio Hoshino (2025) The microcephaly-associated transcriptional regulator AUTS2 cooperates with Polycomb complex PRC2 to produce upper-layer neurons in mice. The EMBO journal
- Kwon HeungSun, Kim Juhyun, Zhou Lecong, Dean Ann (2025) LDB1 regulates gene expression and chromatin structure in pluripotency and lineage differentiation bioRxiv
- Morao Ana Karina, Chervova Almira, Zhao Yuya, Ercan Sevinc, Cecere Germano (2025) DNA supercoiling modulates eukaryotic transcription in a gene-orientation dependent manner bioRxiv
- Aanchal Choudhary, Moonia Ammari, Hyuk Sung Yoon, Mark Zander (2024) High-throughput capture of transcription factor-driven epigenome dynamics using PHILO ChIP-seq Nucleic Acids Research
- Ahmad Kami, Wooten Matt, Takushi Brittany N, Vidaurre Velinda, Chen Xin, Henikoff Steven (2024) Histone H4 limits transcription of the histone locus in Drosophila bioRxiv
- Catalog Number
15-1017-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

