CUTANA™ ChIC/CUT&RUN Kit
For Targeted Release of Genomic Fragments into Solution
Product Description
ChIC and CUT&RUN have revolutionized the study of chromatin regulation by enabling targeted release of genomic fragments into solution. CUTANA™ CUT&RUN kits, reagents, and assay services map histone PTMs and chromatin-interacting proteins with high resolution, at a fraction of the time and cost of standard ChIP-seq experiments.
- Dramatically reduced background
- High resolution genomic mapping for histone PTMs and chromatin-associated proteins
- Small number of cells and only 3-8 million sequencing reads per sample needed
- Streamlined and cost saving workflow
Kit Description
The CUTANA™ ChIC/CUT&RUN Kit enables streamlined chromatin profiling of histone post-translational modifications (PTMs) and chromatin associated proteins while providing increased throughput and reproducibility with multi-channel pipetting. Positive (H3K4me3) and negative (IgG) control antibodies are included to pair with SNAP-CUTANA™ spike-in controls for assay optimization and continuous monitoring (Figure 1). E. coli DNA is included for data normalization. The kit is compatible with a variety of inputs including cells or nuclei derived from native, cryopreserved, or cross-linked samples. While it is recommended to start with 500,000 cells, comparable data can be generated using as few as 5,000 cells. The inclusion of controls, as well as compatibility with diverse target types, sample inputs, and low cell numbers, make this kit the go-to solution for chromatin mapping experiments.
DNA for use in this kit can be easily purified with the CUTANA™ DNA Purification Kit.
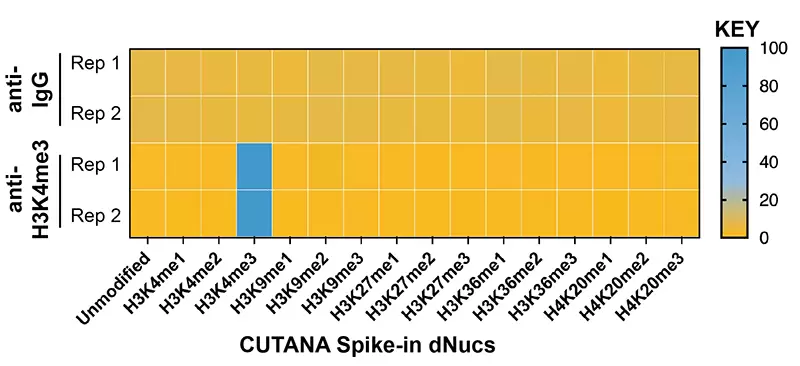
Figure 1: SNAP-CUTANA™ K-MetStat Spike-in Controls. DNA-barcoded designer nucleosomes (dNucs) representing 16 different K-methyl PTM states: mono-, di-, and tri-methylation at H3K4, H3K9, H3K27, H3K36, and H4K20, as well as unmodified control, were spiked into CUT&RUN samples prior to the addition of the control antibodies provided with the kit (IgG, H3K4me3). Instances of each spike-in barcode were counted and normalized from raw fastq files. Barcodes for IgG (top; normalized to the sum of total reads) and H3K4me3 (bottom; normalized to on-target) antibodies provided with this kit are shown. The spike-ins confirmed optimal experimental conditions (H3K4me3 antibody specifically recovered the target dNuc, while IgG showed no preferential enrichment).
Validation Data
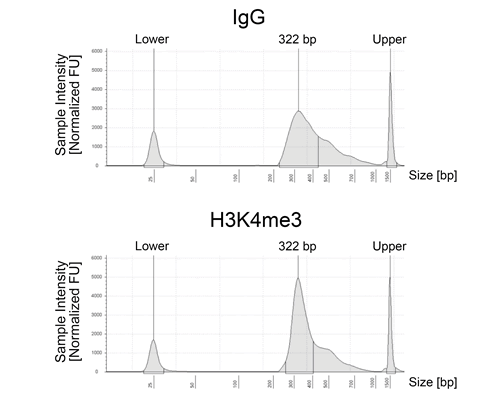
Fig.2. CUT&RUN DNA Fragment Size Distribution Analysis. CUT&RUN was performed using the CUTANA ChIC/CUT&RUN Kit starting with 500,000 K562 cells. CUT&RUN DNA isolated from IgG Negative Control (13-0042k) and H3K4me3 Positive Control (13-0041k) antibodies was used to prepare paired-end Illumina sequencing libraries. Library DNA was analyzed by Agilent Tapestation®. This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN (~300 bp peak represents 150 bp nucleosomes + sequencing adapters).
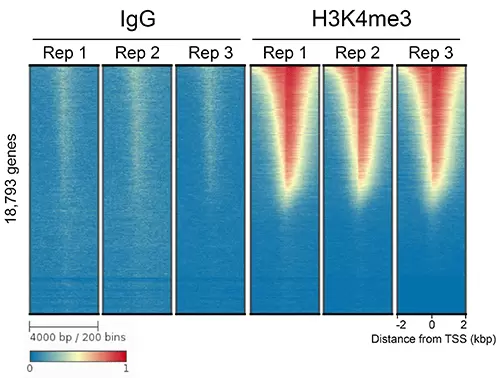
Figure 3: CUT&RUN genome-wide heatmaps. CUT&RUN data was generated using the CUTANA ChIC/CUT&RUN Kit with 500,000 K-562 cells. Three replicates (“Rep”) of IgG (left) and H3K4me3 (right) antibodies are shown in a heatmap. CUT&RUN signal (from an average of 5.9 million paired-end reads) aligned to the transcription start site (TSS, +/- 2kb) are presented for 18,793 genes. High and low signal are ranked by intensity (top to bottom) and reflected by red and blue colors, respectively. Gene rows in each heatmap are aligned and sorted.
Storage and Stability
DO NOT FREEZE ENTIRE KIT. Upon receipt, store individual components at room temperature, 4°C and -20°C (see manual for full instructions).
Related Products
CUTANA CUT&RUN Library Prep Kit with Multiplexing Primer Set 1
CUTANA CUT&RUN Library Prep Kit with Multiplexing Primer Set 2
CUTANA pAG-MNase for ChIC/CUT&RUN Workflows
CUTANA CUT&RUN Compatible Antibodies
Product Citations
- Alireza Lorzadeh, George Ye, Sweta Sharma, Unmesh Jadhav (2025) Motif distribution and DNA methylation underlie distinct Cdx2 binding during development and homeostasis. Nature communications
- Andrea Berardi, Charlotte Leonie Kaestner, Michela Ghitti, Giacomo Quilici, Paolo Cocomazzi, Jianping Li, Federico Ballabio, Chiara Zucchelli, Stefan Knapp, Jonathan D Licht, Giovanna Musco (2025) The C-terminal PHD V C5HCH tandem domain of NSD2 is a combinatorial reader of unmodified H3K4 and tri-methylated H3K27 that regulates transcription of cell adhesion genes in multiple myeloma Nucleic Acids Research
- Chong Wang, Merrin Manlong Leong, Weiyue Ding, Yohei Narita, Xiang Liu, Hongbo Wang, Stefanie P T Yiu, Jessica Lee, Katelyn R S Zhao, Amy Cui, Benjamin Gewurz, Wolfgang Hammerschmidt, Mingxiang Teng, Bo Zhao (2025) Viral oncogene EBNALP regulates YY1 DNA binding and alters host 3D genome organization. EMBO reports
- Jiaying Chen, Na Liu, Hongying Qi, Nils Neuenkirchen, Yuedong Huang, Haifan Lin (2025) Piwi regulates the usage of alternative transcription start sites in the Drosophila ovary Nucleic Acids Research
- Madhavi Murali, Abbas Saeed, Sohyoung Kim, Kyunghee Burkitt, Hui Cheng, Arfa Moshiri, Jawad Akhtar, Daniel Tsai, Marie Luff, Baktiar Karim, Vassiliki Saloura (2025) SMYD3 drives cell cycle and epithelial-mesenchymal transition pathways through dual gene transcriptional repression and activation in HPV-negative head and neck cancer Scientific Reports
- Catalog Number
14-1048-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

