SNAP-ChIP K-MetStat Panel
Product Description
SNAP-ChIP (Sample Normalization and Antibody Profiling for Chromatin Immunoprecipitation) is a groundbreaking new technology that allows you to normalize your ChIP and ChIP-seq experiments as well as profile the specificity of your ChIP antibodies. SNAP-ChIP uses DNA barcoded recombinant designer nucleosomes (dNucs) as next-generation spike-ins for Chromatin Immunoprecipitation (ChIP). EpiCypher's K-MetStat panel can easily be added to any ChIP workflow without altering the protocol. However, users can monitor antibody specificity and evaluate technical variability within a ChIP or ChIP-seq experiment for the first time, setting SNAP-ChIP apart from any other exogenous spike-in controls (i.e. Drosophila) currently available, which do not assess antibody specificity or efficiency and are heterogenous & minimally reproducible.
The SNAP-ChIP K-MetStat Panel includes 1 unmodified nucleosome plus 15 modified nucleosomes with histone H3 or H4 posttranslational modifications (PTMs, created by a proprietary semi-synthetic method): H3K4me1, H3K4me2, H3K4me3, H3K9me1, H3K9me2, H3K9me3, H3K27me1, H3K27me2, H3K27me3, H3K36me1, H3K36me2, H3K36me3, H4K20me1, H4K20me2 and H4K20me3.
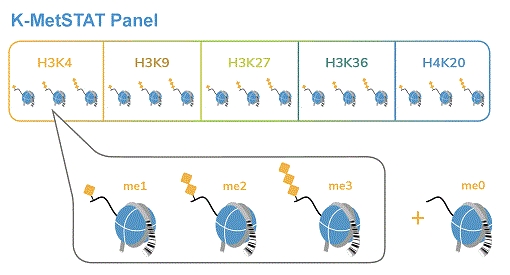
The panel consists of 16 pooled DNA-barcoded recombinant nucleosomes -
1 unmodified and 15 with lysine methyl marks on H3 and H4
SNAP-ChIP Data profiling antibody specificity:
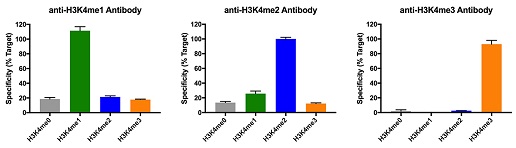
Representative images for SNAP-ChIP K-MetStats (unmodified, H3K4me1, H3K4me2, H3K4me3) assayed in a chromatin immunoprecipitation (ChIP) experiment using commercially available ChIP grade antibodies (3 µg, n = 3). Quantitative Real-Time PCR (qPCR) for the DNA barcodes corresponding to unmodified (H3K4me0), H3K4me1, H3K4me2, and H3K4me3 nucleosomes show recovery of the barcodes corresponding to the expected antibody target. Identical experiments were performed for the remainder of the K-MetStat Panel (H3K9me1, H3K9me2, H3K9me3, H3K27me1, H3K27me2, H3K27me3, H3K36me1, H3K36me2, H3K36me3, H4K20me1, H4K20me2 and H4K20me3).
SNAP-ChIP is extremely useful for determining antibody cross-reactivity with off-target modifications. The cross-reactivity observed above (K4me1 & K4me2) is not indicative of a failure in the SNAP-ChIP procedure, but in the antibody to specifically recognize its target.
Product Citations
- Samantha Carrothers, Rafael Trevisan, Nishad Jayasundara, Nicole Pelletier, Emma Weeks, Joel N. Meyer, Richard Di Giulio, Caren Weinhouse (2025) An epigenetic memory at the CYP1A gene in cancer-resistant, pollution-adapted killifish Scientific Reports
- Segert Julian A., Bulyk Martha L. (2025) Histone H4 lysine 20 monomethylation is not a mark of transcriptional silencers bioRxiv
- Angélica Torres-BerrÃo, Molly Estill, Vishwendra Patel, Aarthi Ramakrishnan, Hope Kronman, Angélica Minier-Toribio, Orna Issler, Caleb J Browne, Eric M Parise, Yentl Y van der Zee, Deena M Walker, Freddyson J MartÃnez-Rivera, Casey K Lardner, Romain Durand-de Cuttoli, Scott J Russo, Li Shen, Simone Sidoli, Eric J Nestler (2024) Mono-methylation of lysine 27 at histone 3 confers lifelong susceptibility to stress. Neuron
- Jeffrey K. Bailey, Dzwokai Ma, Dennis O. Clegg, Yusuke Kamachi (2024) Initial Characterization of WDR5B Reveals a Role in the Proliferation of Retinal Pigment Epithelial Cells Cells
- Li Peilin, Fujisawa Tatsuya, Narita Haruka, Uneme Yuta, Seki Masahide, Hu Mengwen, Narumi Ryohei, Namekawa Satoshi H., Khan Shehroz S., Davie James R., Suzuki Yutaka, Adachi Jun, Ashraf Ahmed, Inoue Azusa, Siomi Mikiko C., Yamanaka Soichiro (2024) Priming Epigenetic Landscape at Gene Promoters through Transcriptional Activation in Mammalian Germ Cells bioRxiv
References
- Shah RN et al. (2018) Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies Molecular Cell
- Catalog Number
19-1001-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

