
SNAP-ChIP Panels of Barcoded Recombinant Nucleosomes
Sample normalization and antibody profiling for chromatin immunoprecipitation
- Mix of homogenous defined DNA-barcoded human recombinant nucleosomes subjected to rigorous quality control for lot-to-lot consistency
- Defined spike-in controls for ChIP and ChIP-Seq reactions
- Useful for ChIP Antibody validation and profiling
- Determine ChIP antibody specificity and target pulldown efficiency
- Monitor technical variability within experiments
- Quantitative recovery of DNA barcodes via qPCR provides useful STOP / GO
capability before advancing to NGS - Perform consistent effector protein binding experiments
SNAP-ChIP (Sample Normalization and Antibody Profiling Chromatin ImmunoPrecipitation) uses DNA-barcoded designer nucleosomes (dNucs) bearing distinct post-translational modifications (PTMs) as nextgeneration spike-in controls for ChIP. EpiCypher’s SNAP-ChIP panels are directly compatible with your current ChIP workflow, with semi-synthetic nucleosomes bearing the PTM of interest immunoprecipitated and processed alongside sample chromatin. Recovery of the associated DNA barcodes is deciphered by quantitative PCR (ChIPqPCR) or Next-Generation Sequencing (ChIP-seq). SNAP-ChIP provides defined standards to evaluate antibody performance and monitor technical variability in ChIP, setting it apart from other spike-in controls.
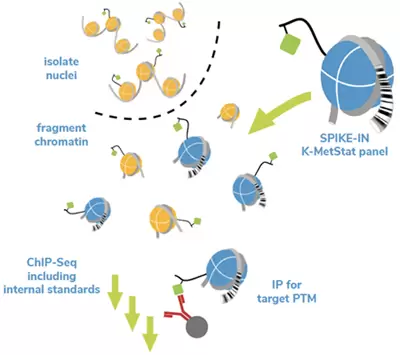
Figure 1. Overview of SNAP-ChIP (adapted from ICeChIP technology). A pool of recombinant dNucs with defined PTMs identified by unique DNA barcodes is added to sample chromatin prior to immunoprecipitation (IP). Capture of the barcoded nucleosomes (on- and off-target) allows the user to assess antibody performance (specificity and target enrichment) and monitor technical variability.
SNAP-ChIP panels are a pool of distinctly modified mononucleosomes assembled from recombinant human histones expressed in E. coli (two each of histones H2A, H2B, H3.3 and H4) wrapped by 147 base pairs of barcoded Widom 601 positioning sequence DNA. Each distinctly modified nucleosome is distinguishable by a unique sequence of DNA (“barcode”) at the 3’ end that can be deciphered by qPCR or next-generation sequencing. Each of the nucleosomes in the pool is wrapped by 2 DNA species allowing for an internal technical replicate.
Available focused sets
The SNAP-ChIP K-MetStat Panel includes 1 unmodified nucleosome plus 15 modified nucleosomes with histone H3 or H4 posttranslational modifications (PTMs, created by a proprietary semi-synthetic method).
The SNAP-ChIP K-AcylStat Panel includes 1 unmodified nucleosome plus 22 modified nucleosomes with histone H2A, H3, or H4 posttranslational modifications (PTMs, created by a proprietary semi-synthetic method).
The SNAP-ChIP OncoStat Panel includes 1 unmodified nucleosome plus 7 modified nucleosomes with histone H3.3 point mutations.