
RNA Methylation Analysis
RNA Methylation Analysis
Comprehensive range of products for analysis of m6A and 5-mC RNA
RNA methylation occurs in different RNA species including tRNA, rRNA, mRNA, tmRNA, snRNA, snoRNA, miRNA, and viral RNA. Different catalytic strategies are employed for RNA methylation by a variety of RNA-methyltransferases. As a post-translational modification, RNA methylation plays a significant role as an epigenetic mechanism.
N6-methyladenosine (m6A) is the most common and abundant methylation modification in RNA molecules present in eukaryotes. 5-methylcytosine (5-mC) also commonly occurs in various RNA molecules.
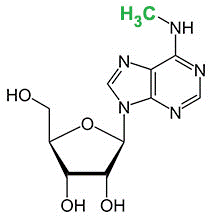
N6-methyladenosine (m6A)
Recent data strongly suggest that m6A and 5-mC RNA methylation affects the regulation of various biological processes such as RNA stability and mRNA translation, and that abnormal RNA methylation contributes to etiology of human diseases. Furthermore, loss of 5-mC in vault RNAs causes aberrant processing into Argonaute-associated small RNA fragments that can function as microRNAs. Researchers continue to investigate m6A and 5-mC in RNA to uncover the mysteries of RNA methylation.
Global m6A & 5-mC RNA Quantification
EpigenTek's m6A and 5-mC RNA methylation kits make it easy for researchers to determine how RNA methylation influences their research project. The kits allow for the direct quantification of either N6-Methyladenosine (m6A) or measuring global 5-mC RNA using total RNA isolated from any species, including mammals, plants, fungi, bacteria, and viruses.
RNA m6A Methylase and Demethylase Activity
For measuring total m6A demethylase activity/inhibition using nuclear extracts or purified m6A demethylases like FTO and ALKBH5 from a broad range of species.
RNA Bisulfite Conversion & cDNA Synthesis
Bisulfite treatment techniques, also known as bisulfite conversion, are the most ideal approaches for preparing RNA for site-specific methylation analysis. This process converts cytosine into uracil, leaving methylated cytosine intact which is then easily distinguishable via high-throughput sequencing. Bisulfite converted RNA can also be used to generate cDNA for use in various downstream applications to analyze methylation condition. Besides the Bisulfit Conversion Kit and the cDNA Synthesis Kit EpigenTek also offers a complete kit for RNA bisulfite conversion and NGS library preparation for 5-methylcytosine RNA-seq.
Methylated RNA Immunoprecipiation (MeRIP)
Get high resolution mapping of m6A RNA using EpigenTek's EpiQuik CUT&RUN m6A RNA (MeRIP) Enrichment Kit and EpiNext CUT&RUN RNA m6A-Seq Kit. With a unique nucleic acid cleavage enzyme mix, these kits isolate only RNA segments at the antibody binding locations and simultaneously cleave/remove the segments of RNA sequence just under the target area. Both our kits can enrich RNA fragments containing the m6A modification from low input RNA, with minimal non-specific background levels. The CUT&RUN RNA m6A-Seq Kit includes the library preparation step as an added benefit.
The Methylated RNA Enrichment and Quantification Kits from RayBiotech identify the abundance and enrichment location of m1A, m6A or7G by MeRIP and quantitative real-time PCR.
The EpiNext CUT&LUNCH RNA Immunoprecipitation (RIP) Kit is designed for ultra-fast, highly specific enrichment of target protein-RNA complexes.
Antibodies to Study RNA Methylation
N6-methyladenosine (m6A) is one of the most common and abundant modifications found in RNA molecules present in eukaryotes. The chemical properties of m6A make it challenging to distinguish; therefore, methods to analyze this modification are highly dependent upon the quality of the ant-m6A antibody used. Both our polyclonal and monoclonal m6A antibodies can be reliably used for the specific detection of m6A in RNA. They have been thoroughly tested and are guaranteed to work in their respective applications.