EpiNext CUT&RUN RNA m6A-Seq Kit
For all-in-one high enrichment meRIP and library preparation for m6A sequencing
Product Description
The EpiNext™ CUT&RUN RNA m6A-Seq Kit is a complete set of optimized buffers and reagents designed to enrich an RNA fragment containing m6A from low input RNA and to prepare a library for next generation sequencing using Illumina platforms such as Illumina Genome Analyzer II, HiSeq and MiSeq systems.
The enriched RNA is specifically suitable to construct both non-barcoded (singleplexed) and barcoded (multiplexed) libraries quickly, allowing m6A regions to be mapped with less bias and at a high resolution.
- High enrichment of RNA fragments containing m6A
- Low Input RNA
- Minimal Background
- Fast, streamlined procedure
- Highly convenient
Background Information
N6-methyl-adenosine (m6A) is the most common and abundant modification on RNA molecules present in eukaryotes. The m6A modification is catalyzed by a methyltransferase complex METTL3 and removed by m6A RNA demethylases FTO and ALKBH5, which catalyze m6A demethylation in an α-ketoglutarate (α-KG)- and Fe2+-dependent manner. METTL3, FTO, and ALKBH5 are known to play important roles in many biological processes, ranging from development and metabolism to fertility.
m6A accounts for more than 80% of all RNA base methylations and exists in various species. m6A is mainly distributed in mRNA and also occurs in non-coding RNA, such as tRNA, rRNA, and snRNA.
The relative abundance of m6A in mRNA transcripts has been shown to affect RNA metabolism processes such as splicing, nuclear export, translation ability and stability, and RNA transcription. Abnormal m6A methylation levels induced by defects in m6A RNA methylase and demethylase could lead to dysfunction of RNA and cause disease. For example, abnormally low levels of m6A in target mRNAs due to increased FTO activity in patients with FTO mutations, through an as-yet-undefined pathway, contributes to the onset of obesity and related diseases.
The dynamic and reversible chemical m6A modification on RNA may also serve as a novel epigenetic marker of profound biological significance. Therefore, more useful information for a better understanding of m6A RNA methylation levels and distribution on RNA transcripts could benefit diagnostics and therapeutics of disease.

Fig 1. Workflow of the EpiNext™ CUT&RUN RNA m6A-Seq Kit.
Principle & Procedure
This kit contains all necessary reagents required for carrying out a successful m6A-Seq starting from total RNA. In the reaction, RNA sequences at both ends of the target m6A-containing regions are cleaved/removed and the target m6A-containing fragments are pulled down using a beads-bound m6A capture antibody.
The target m6A-containing RNA fragments are then reverse-transcripted ligated with adaptors. The ligated RNA is released, purified, and amplified with a high-fidelity PCR mix for library cDNA construction.
The cDNA is then cleaned, released, and eluted. Included in the kit is a negative control non-immune IgG, which can demonstrate the efficacy of the kit and performance at the enriched RNA quantification or bioanalyzer analysis step.
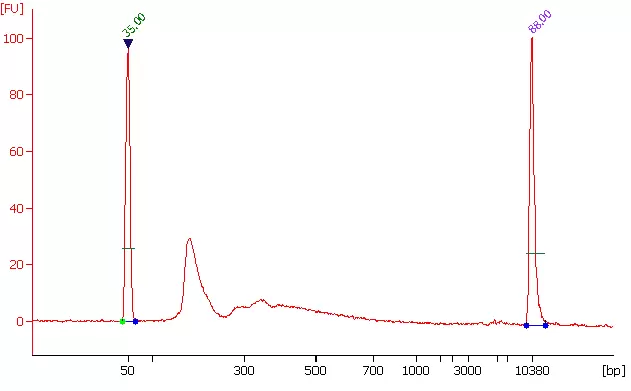
Fig 2. Size distribution of library fragments. m6A fragments were enriched from 3 ug of total RNA by the anti- m6A antibody (EpigenTek, #A-1801) and used for cDNA library preparation. Peak at 195 bps reflects insert size (around 55 bps) of RNA bound by m6A antibody.
Product Citations
- Kumar RP, Kumar R, Ganguly A, Ghosh A, Ray S, Islam MR, Saha A, Roy N, Dasgupta P, Knowles T, Niloy AJ, Marsh C, Paul S (2024) METTL3 shapes m6A epitranscriptomic landscape for successful human placentation. bioRxiv
- Shiraishi C, Matsumoto A, Ichihara K, Yamamoto T, Yokoyama T, Mizoo T, Hatano A, Matsumoto M, Tanaka Y, Matsuura-Suzuki E, Iwasaki S, Matsushima S, Tsutsui H, Nakayama KI (2023) RPL3L-containing ribosomes determine translation elongation dynamics required for cardiac function. Nat Commun
- Shabani D, Dresselhaus T, Dukowic-Schulze S (2022) Profiling m6A RNA Modifications in Low Amounts of Plant Cells Using Maize Meiocytes. Methods Mol Biol

