HA Tag CUTANA™ CUT&RUN Antibody
Useful for studies utilizing HA-tagged target proteins
Product Description
This antibody meets EpiCypher’s "CUTANA Compatible" criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping. Every lot of a CUTANA Compatible antibody is tested in the indicated approach using EpiCypher optimized protocols and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target. HA antibody is useful for studies utilizing HA-tagged target proteins. HA Tag antibody produces CUT&RUN peaks (Figure 1) that overlap with GATA3 DNA-binding motifs (Figure 2) in breast cancer cells expressing 3xHA-tagged GATA3 transcription factor [Takaku et al., 2016]*
*Thanks to Dr. Takaku (UND) for 3xFlag-GATA3-3xHA MDA-MB-231 cells.
Related Product
Pair with our SNAP-CUTANA™ HA Tag Panel for CUT&RUN success.
Validation Data
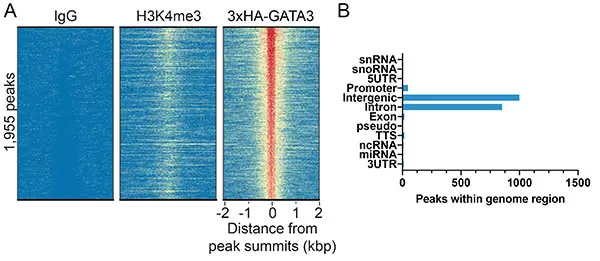
Figure 1: HA-tagged protein peaks in CUT&RUN. CUT&RUN was performed using HA antibody (0.5 µg) with 500,000 MDA-MB-231 cells stably expressing 3xHA-tagged GATA3 [1]. Peaks were called using MACS2. (A) Heatmap showing 3xHA-GATA3 peaks relative to IgG and H3K4me3 control antibodies (Cat.No. 13-0042-EPC and 13-0041-EPC, respectively) in aligned rows ranked by intensity (top to bottom) and colored such that red indicates high localized enrichment and blue denotes background signal. (B) The number of 3xHA-GATA3 peaks which fall into distinct classes of functionally annotated genomic regions is plotted.

Figure 2: HA-tagged transcription factor binding motif analysis in CUT&RUN. (A) Homer analysis determined that the GATA3 consensus motif, represented as a sequence logo position weight matrix, was enriched under GATA3-3xHA CUT&RUN peaks. (B) The number of GATA3-3xHA peaks containing GATA3 consensus motifs from panel A is represented by a Venn Diagram. (C-D) Two representative loci show overlap of GATA3-3xHA peaks with the consensus motifs noted by tick marks beneath the tracks.

Figure 3: Target-specific epitope cleavage of HA Tag antibody in CUT&RUN was determined using DNA-barcoded recombinant nucleosome spike-in controls.
(A) A panel of recombinant nucleosomes was created where various epitope tags (3xTY1, 3xFLAG, 3xHA) were fused to the histone H3 tail. The fused nucleosomes and an unmodified control were immobilized to streptavidin beads (SA Bead) and spiked into CUT&RUN samples alongside ConA bead immobilized MDA-MB-231 cells expressing GATA3-3xHA (Figure 1). HA Tag antibody and pAG-MNase (Cat.No. 15-1016-EPC) were then added to release antibody-bound nucleosomes into solution through pAG-MNase mediated cleavage of the linker DNA (light blue). This approach provided a defined experimental control to assess whether the HA Tag antibody selectively cleaved the target epitope with high specificity and minimal background. (B) CUT&RUN sequence reads were aligned to the unique DNA "barcodes" corresponding to each nucleosome in the spike-in panel. Data are expressed as the percent of reads recovered relative to the intended target (3xHA, set to 100%). This analysis confirms that the HA Tag antibody specifically liberated the target epitope-tagged nucleosome into solution.
Product Citations
- Cui Jieying H., Kwan James Z.J., Faghihi Armin, Nguyen Thomas F., Teves Sheila S. (2024) Functional divergence of TBP homologs through distinct DNA binding dynamics bioRxiv
- Sedona Eve Murphy, Alistair Nicol Boettiger (2024) Polycomb repression of Hox genes involves spatial feedback but not domain compaction or phase transition. Nature genetics
- Vandeuren Adrianna L., O’Dare Kierney, Wilson Rosemary H. C., van Eijk Patrick, Julio Lindsay R., MacLeod Shannon G., Chee Ella, Salpukas Annika, Kriz Emma M., Lantz George A., Gordon Shellaina, Elsässer Simon J., Reed Simon H., Day Tovah A. (2024) The Role of BAZ2-dependent Chromatin Remodeling in Suppressing G4 DNA Structures and Associated Genomic Instability bioRxiv
- Zian Liao, Suni Tang, Kaori Nozawa, Keisuke Shimada, Masahito Ikawa, Diana Monsivais, Martin Matzuk, Carmen J Williams, Wei Yan (2024) Affinity-tagged SMAD1 and SMAD5 mouse lines reveal transcriptional reprogramming mechanisms during early pregnancy eLife
- Ahmed H. Ghobashi, Truc T. Vuong, Jane W. Kimani, Christopher A. Ladaika, Peter C. Hollenhorst, Heather M. O’Hagan (2023) Activation of AKT induces EZH2-mediated β-catenin trimethylation in colorectal cancer iScience
References
- Takaku et al. (2016) GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler Genome Biol.
- Catalog Number
13-2010-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

