MethylFlash 5-mC RNA Methylation ELISA Easy Kit (Fluorometric)
Product Description
The MethylFlash™ 5-mC RNA Methylation ELISA Easy Kit (Fluorometric) is a complete set of optimized buffers and reagents to fluorometrically quantify global 5-methylcytosine (5-mC) RNA methylation levels using total RNA isolated from any species including mammals, plants, fungi, bacteria, and viruses in a variety of forms including, but not limited to, cultured cells, fresh and frozen tissues, plasma/serum samples and body fluid samples, etc. This kit has the following advantages:
- Entire procedure only needs 2 hours and 40 minutes
- Assay has a large “signal window" with less variation between replicates
- Inherently low background noise, no need for plate blocking steps
- Detection limit can be as low as 0.02% of 5-mC RNA from 200 ng of input RNA
- High specificity to 5-mC
- Positive and negative controls allow detection of 5-mC RNA methylation in any species
- Most accurate and highly comparable with HPLC-MS analysis
- Strip-well microplate format for manual or high throughput analysis
Background Information
5-methylcytosine (5-mC) in DNA occurs by the covalent addition of a methyl group at the 5-carbon of the cytosine ring by DNA methyltransferases. This process has been well studied and is generally associated with repression of gene expression. It was also observed that in humans, 5-mC occurs in various RNA molecules including tRNAs, rRNAs, mRNAs and non-coding RNAs (ncRNAs). 5-mC seems to be enriched in some classes of ncRNA, but relatively depleted in mRNAs. Levels of 5-mC are variable in animal genomes, ranging from undetectable amounts in some insects to about 0.1-0.45% of total RNA in human cells. The majority (83%) of 5-mC sites were found in mRNAs. Within these transcripts 5-mC appears to be depleted within protein coding sequences but enriched in 5’ and 3’ UTRs. Two different methyltransferases, NSUN2 and Dnmt2 are known to catalyze 5-mC modification in eukaryotic RNA. There has been strong evidence that RNA cytosine methylation affects the regulation of various biological processes such as RNA stability and mRNA translation. Furthermore, loss of 5-mC in vault RNAs causes aberrant processing into Argonaute-associated small RNA fragments that can function as microRNAs. Thus, impaired processing of vault ncRNA may contribute to the etiology of human disorders related to NSun2-defciency.
Principle & Procedure
This kit contains all reagents necessary for the quantification of global 5-mC RNA methylation. In the assay, RNA is bound to strip-wells that are specifically treated to have a high nucleic acid affinity. 5-mC in RNA is detected using capture and detection antibodies and then quantified fluorometrically by reading the fluorescence in a microplate spectrophotometer. The percentage of 5-mC RNA is proportional to the fluorescence intensity measured.
Fig. 1. Schematic procedure for the MethylFlash™ 5-mC RNA Methylation ELISA Easy Kit (Fluorometric).
Starting Materials
Input RNA should be highly pure with 260/280 ratio >2.0 and relatively free of DNA. DNase I can be used to remove DNA. RNA should be eluted in RNase-free water. The RNA amount can range from 50 ng to 300 ng per reaction. However, we recommend using 200 ng of RNA, which is the optimized input amount for the best results.
Performance Data
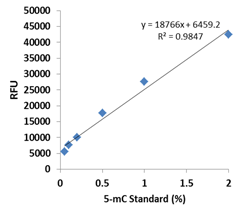
Fig. 2. Example of an optimal standard curve generated with 5-mC standard control.
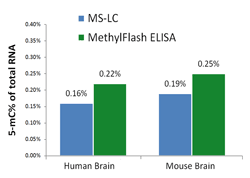
Fig. 3. Accurate quantification of 5-mC content of various RNA samples from different species using the MethylFlash™ 5-mC RNA Methylation ELISA Easy Kit (Fluorometric). The results are closely correlated with those obtained by MS-LC.
Product Citations
- Liu L, Chen Y, Zhang T, Cui G, Wang W, Zhang G, Li J, Zhang Y, Wang Y, Zou Y, Ren Z, Xue W, Sun R (2024) YBX1 Promotes Esophageal Squamous Cell Carcinoma Progression via m5Câ€Dependent SMOX mRNA Stabilization. Adv Sci (Weinh)
- Yu T, Nie FQ, Zhang Q, Yu SK, Zhang ML, Wang Q, Wang EX, Lu KH, Sun M (2024) Effects of methionine deficiency on B7H3-DAP12-CAR-T cells in the treatment of lung squamous cell carcinoma. Cell Death Dis
- Wang Y, Wei J, Feng L, Li O, Huang L, Zhou S, Xu Y, An K, Zhang Y, Chen R, He L, Wang Q, Wang H, Du Y, Liu R, Huang C, Zhang X, Yang YG, Kan Q, Tian X (2023) Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol Cancer
- Zhang G, Liu L, Li J, Chen Y, Wang Y, Zhang Y, Dong Z, Xue W, Sun R, Cui G (2023) NSUN2 stimulates tumor progression via enhancing TIAM2 mRNA stability in pancreatic cancer. Cell Death Discov
- Betlej G, Ząbek T, Lewińska A, Błoniarz D, Rzeszutek I, Wnuk M (2022) RNA 5-methylcytosine status is associated with DNMT2/TRDMT1 nuclear localization in osteosarcoma cell lines. J Bone Oncol

