EpiNext High-Sensitivity Bisulfite-Seq Kit (Illumina)
Designed to prepare bisulfite-converted DNA libraries for Illumina platform-based bisulfite sequencing
Product Description
The EpiNext™ High-Sensitivity Bisulfite-Seq Kit is designed to prepare bisulfite-converted DNA libraries for Illumina platform-based bisulfite sequencing including whole genome bisulfite sequencing, oxidative bisulfite sequencing, reduced representation bisulfite sequencing, and other bisulfite-next generation sequencing.
The optimized protocol and components of the kit allow subnanogram DNA to be used for simultaneous bisulfite conversion and fragmentation followed by non-barcoded (singleplexed) and barcoded (multiplexed) library construction in less than 7 hours.
Features and Advantages:
- Allows for simultaneous bisulfite conversion and size-appropriate DNA fragmentation. The bisulfite DNA can be directly ligated to adaptors thereby eliminating the possibility of breaking adaptor-ligated fragments, which often occurs with currently used WGBS and RRBS methods.
- The procedure from DNA bisulfite treatment to ready-to-use library DNA can be completed within 6 hours and 30 minutes.
- Completely converts unmethylated cytosine into uracil (>99%) with negligible inappropriate- or error-conversions of methylcytosine to thymine.
- High sensitivity and efficiency - Innovative adaptor ligation of bisulfite DNA eliminates loss of fragments and selection bias, which enables input DNA to be as low as 0.2 ng, making it ideal for methylation profiling of precious, limited samples. The kit can be used for both non-barcoded (singleplexed) and barcoded (multiplexed) DNA library preparation.
- The kit contains all required components for each step of DNA library preparation, which are sufficient for bisulfite conversion, ligation, clean-up, size selection and library amplification, thereby allowing the bisulfite DNA library preparation to be streamlined for the most reliable and consistent results.
- Ultra HiFi amplification enables achievement of reproducibly high yields of DNA libraries with minimal sequence bias and low error rates.
- Starting materials can be genomic DNA isolated from various tissue/cell samples such as fresh and frozen tissue, cultured cells from a flask or microplate, microdissection samples, paraffin-embedded tissue, biopsy, embryonic cells, plasma/serum samples, and body fluid samples, etc. DNA enriched from various enrichment reactions such as ChIP, MeDIP/hMeDIP or exon capture may be also used as starting material.
Background Information
DNA methylation occurs by the covalent addition of a methyl group (CH3) at the 5-carbon of the cytosine ring, resulting in 5-methylcytosine (5-mC). DNA methylation is essential in regulating gene expression in nearly all biological processes including development, growth, and differentiation.
Alterations in DNA methylation have been demonstrated to cause a change in gene expression. For example, hypermethylation leads to gene silencing or decreased gene expression while hypomethylation activates genes or increases gene expression. Aberrant DNA methylation is also associated with pathogenesis of diseases such as cancer, autoimmune disorders, and schizophrenia.
Thus, genome-wide analysis of DNA methylation could provide valuable information for discovering epigenetic markers used for disease diagnosis, and potential targets used for therapeutics.
Principle and Procedure
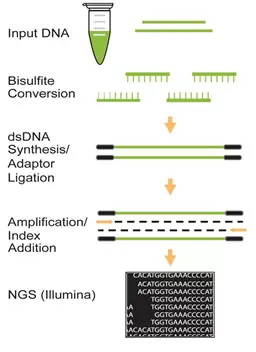
This kit includes all reagents required for successfully preparing a library directly using bisulfite DNA generated from a tiny amount of input DNA. In this preparation, DNA is simultaneously bisulfite converted and fragmented to the appropriate length during the bisulfite process.
The bisulfite-treated DNA, which is in single stranded form, is then simultaneously converted to dsDNA and adaptor ligated. The ligated fragments are size selected and purified using MQ Binding Beads, followed by amplification with a high-fidelity PCR Mix which ensures maximum yields from minimum amounts of starting material and provides highly accurate amplification of library DNA with low error rates and minimum bias.
Starting Materials
Starting materials can be genomic DNA isolated from various tissue/cell samples such as fresh and frozen tissue, cultured cells from a flask or microplate, microdissection samples, FFPE tissue, plasma/serum, and body fluid samples, etc. DNA enriched from various enrichment reactions such as ChIP, MeDIP/hMeDIP or exon capture may also be used as starting material. DNA should be without any previous restriction digestion step. Plasmid DNA can be used for bisulfite treatment with or without previous linearization, as the kit allows for DNA denaturation status to remain during the entire DNA bisulfite conversion process and direct ligation of adaptors to bisulfite DNA. Input amount of DNA can be from 0.2 ng to 200 ng. For optimal preparation, the input amount should be 10-50 ng.
Multiplexing
For constructing multiplex DNA libraries used for next-generation sequencing with the Illumina platform including GAIIx, HiSeq and MiSeq, the EpiNext™ NGS Barcode (Index) Set-12 is available.
Product Citations
- Perez-Maldonado MA, González-González XA, Chimal-Monroy J, MarÃn-Llera JC (2024) Influence of DNA-methylation at multiple stages of limb chondrogenesis. Dev Biol
- Halawa S, Latif N, Tseng YT, Ibrahim AM, Chester AH, Moustafa A, Aguib Y, Yacoub MH (2022) Profiling Genome-Wide DNA Methylation Patterns in Human Aortic and Mitral Valves. Front Cardiovasc Med
- Shpokayte M, McKissick O, Guan X, Yuan B, Rahsepar B, Fernandez FR, Ruesch E, Grella SL, White JA, Liu XS, Ramirez S (2022) Hippocampal cells segregate positive and negative engrams. Commun Biol
- Buenahora MR, Lafaurie GI, Perdomo SJ (2020) Identification of HPV16-p16INK4abeta mediated methylation in oral potentially malignant disorder. Epigenetics
- Li Y, Tríbulo P, Bakhtiarizadeh MR, Siqueira LG, Ji T, Rivera RM, Hansen PJ (2019) Conditions of embryo culture from days 5 to 7 of development alter the DNA methylome of the bovine fetus at day 86 of gestation. J Assist Reprod Genet

