miND® Spike-In (resuspended)
Spike-Ins for small RNA-sequencing quality control and absolute normalization
Product Description
miND® Spike-Ins are a proprietary oligonucleotide mix with unique design features (Lutzmayer et al.) added to your total RNA samples prior to microRNA or small RNA next-generation sequencing analysis.
Our partner TAmiRNA has optimized the sequence composition and concentration range to be compatible with almost any species as well as a broad range of sample types including biofluids (serum, plasma, urine, CSF, synovial fluid), cells, tissues, extracellular vesicles, and non-vesicular fractions (Khamina et al.).
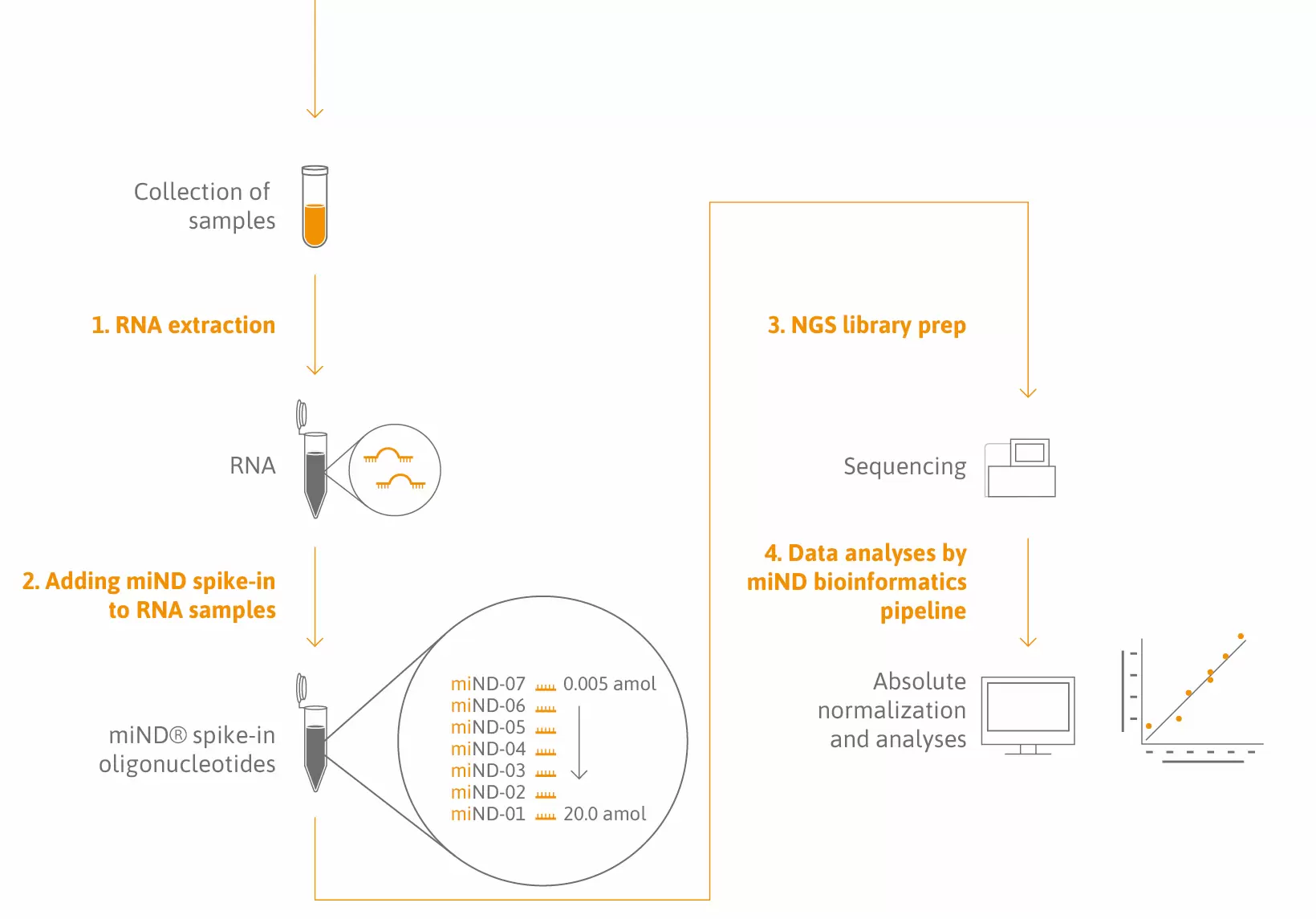
miND® Spike-Ins fulfill two main purposes:
1. Quality control: confirm the high dynamic range and quantitativeness of your small RNA-sequencing experiments.
2. Absolute normalization: convert read counts to copies/µl total RNA and reduce bias originating from variation in RNA composition between samples.
Benefits
The challenge: small RNA-sequencing, especially for challenging input samples such as biofluids, exosomes, or low cell numbers, is prone to sequencing bias. In addition, the comparison of microRNAs and other small RNAs between sample types with differing RNA compositions is skewed based on the assumption of a constant amount of small RNAs per sample (the underlying hypothesis for reads per million (RPM) normalization).
The solution: miND® Spike-Ins are a novel quality control parameter and normalizer that consists of seven oligonucleotides, each characterized by a unique core sequence flanked by 4 randomized nucleotides. miND® Spike-Ins are provided in a specific ratio to cover the broad concentration range of endogenous small RNAs.

Simplicity: miND® Spike-Ins come as ready-to-use reagents. No dilution, mixing, or change in your workflow is required – miND® spike-ins are directly added to your RNA sample and used for NGS library preparation.
Sample-to-insight: A public data analysis pipeline was specifically developed to convert small RNA-sequencing raw data into a simple but comprehensive report providing full access to your data in the conventional way (RPM, read count) as well as in absolute concentrations. On top of that, TAmiRNA has added several unsupervised and supervised analyses and QC parameters to the report.
How does it work?
- 7 unique core sequences mixed in a specific proportions to cover a wide range of endogenous microRNAs
- Each unique core sequence consists of 13 nt that doesn’t match the genome of interest
- The core sequence is flanked by a set of 4 randomized nucleotides on both 5’ and 3’ ends
- Each of these 13 nt unique core sequences can be represented by up to 65,536 possible 21 nt sequences
- The miND spike-in contains up to 458,752 unique oligonucleotides
Product Citations
- Anderson JR., Jacobsen S., Walters M. et al. (2022) Small non-coding RNA landscape of extracellular vesicles from a post-traumatic model of equine osteoarthritis Front. Vet. Sci. Sec. Veterinary Regenerative Medicine
- Diendorfer A, Khamina K, Pultar M and Hackl M (2022) miND (miRNA NGS Discovery pipeline): a small RNA-seq analysis pipeline and report generator for microRNA biomarker discovery studies F1000Research, 11:233, 122
- Khamina K, Diendorfer A, Skalicky S, et al. (2022) A MicroRNA Next-Generation-Sequencing Discovery Assay (miND) for Genome-Scale Analysis and Absolute Quantitation of Circulating MicroRNA Biomarkers Int. J. Mol. Sc . 23(3), 1226
- Lutzmayer S, Enugutti B and Nodine MD (2017) Novel small RNA spike-in oligonucleotides enable absolute normalization of small RNA-Seq data Sci Rep.7(1):5913. doi: 10.1038/s41598-017-06174-3.
- Catalog Number
KT-041-MIND-48-TAM - Supplier
TamiRNA - Size
- Shipping
Dry Ice

