RealSeq-AC Kit for small RNA Library Construction from total RNA samples from tissue or cells (Illumina Platform)
Reducing Bias in Small-RNA Sequencing
Product Description
- Accurately quantifies biologically relevant small RNAs
- Eliminates bias-induced over/under represented small RNAs
- Allows discovery of novel small RNAs
- Inputs between 1 ng to 100 ng of total RNA
Workflow
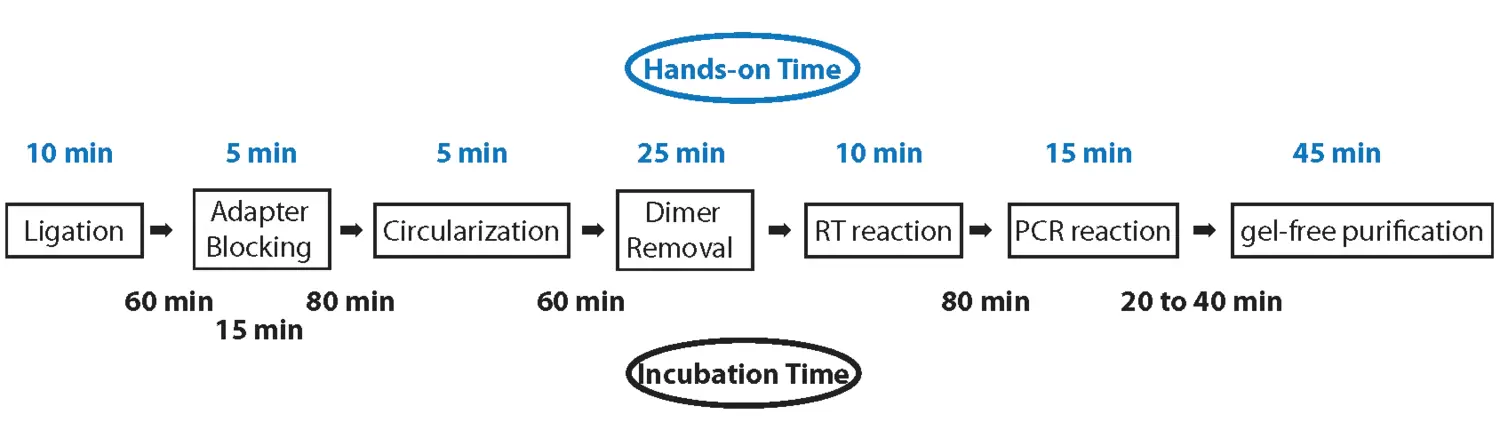
3´ ends of miRNAs or any other 5’-p and 3’-OH small RNAs are ligated to RealSeq® adapters which contain standard adapter sequences (shown in blue and green) that are compatible with Illumina/Solexa sequencing.
Unligated adapters are blocked before circularization of the miRNA-adapter product and then removed. This intramolecular ligation is significantly more efficient than intermolecular ligation of 5’-adapters in standard two-adapter ligation schemes.
Reverse transcription of the circularized miRNA-adapter products yields monomer cDNAs with the targets sequences flanked by Illumina 5’- and 3’-adapter sequences. PCR amplification of the RT product with the extended, bar-coded PCR primers, yields amplicons ready for size selection using SPRI select technology (Beckman Coulter) which is included in RealSeq®-AC and RealSeq®-biofluids kits.
Related Products
Additional Reverse Index Primers
Selective Suppression Probes (SSP) for RNA Depletion - Human Plasma Panel
Custom Selective Suppression Probes (SSPs) for RNA Depletion
Supporting Data
Reducing bias in small-RNA sequencing
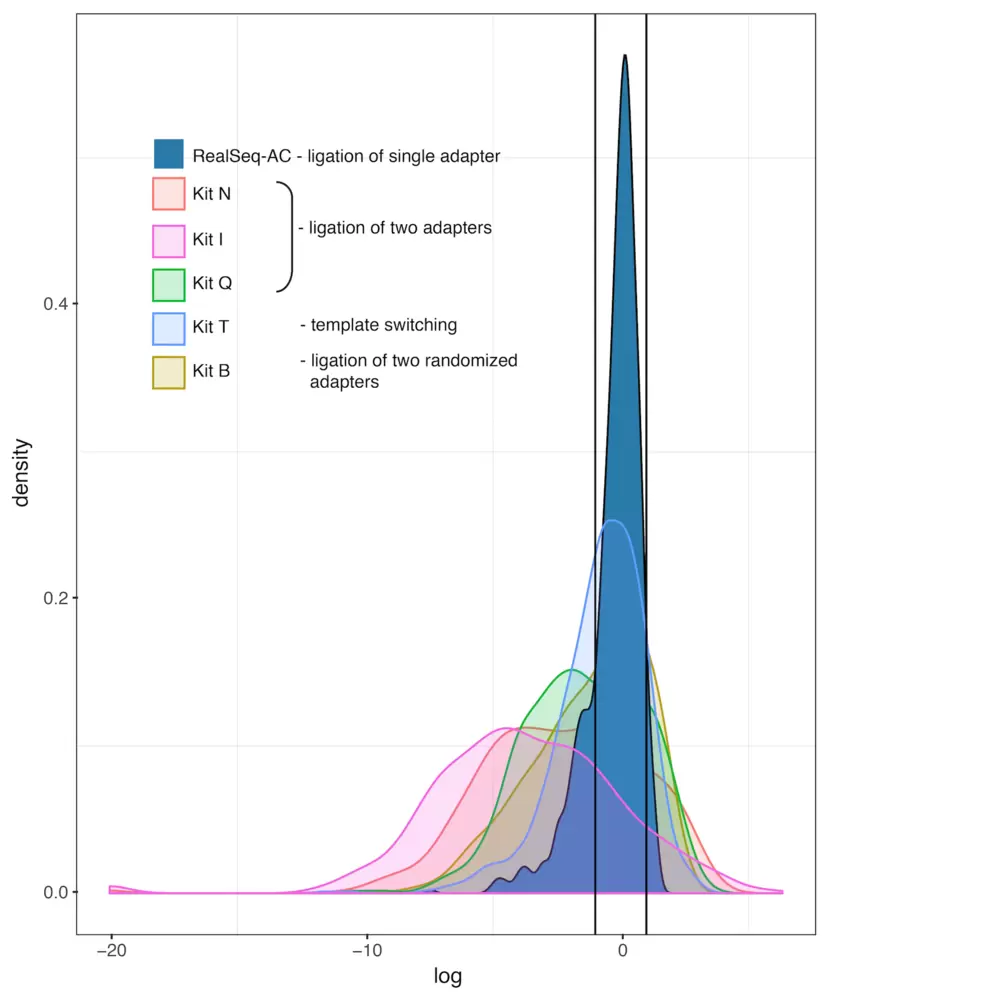
Fig.1. One pmole of miRXPlore Universal Reference pool (Miltenyi Biotec) was used to compare incorporation bias in six different commercially available library preparation kits. Purified libraries were sequenced on the Illumina MiSeq platform. Trimmed sequencing reads were aligned to a custom miRNA reference. Reads mapping to miRNAs were counted and fold-deviations from the equimolar input were calculated and plotted as log2 values. Measurements of miRNA levels within a factor of two of the expected values (between vertical lines) are considered unbiased (Fuchs et al, 2015). The method of adapter attachment to the miRNAs is noted in the legend.
RealSeq®-AC is highly efficient, detecting more miRNAs in total RNA samples
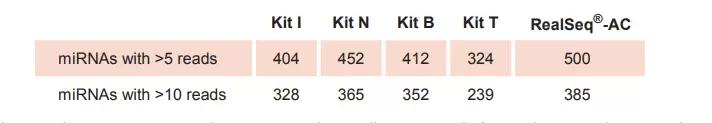
Fig.2. RealSeq®-AC detects more miRNAs with over 5 or 10 reads per million, respectively, from total RNA samples compared to other kits for miRNA sequencing library preparation. RealSeq®-AC is optimized for inputs between 1 ng to 100 ng of total RNA. Lower numbers of PCR cycles are reducing PCR-induced issue.
Product Citations
- Hamdi, M., et al. (2024) Oviductal extracellular vesicles miRNA cargo varies in response to embryos and their quality BMC Genomics
- Androvic, P., et al. (2022) Small RNA-Sequencing for Analysis of Circulating miRNAs: Benchmark Study J. Mol. Diagn.
- Carro Vázquez, D. et al. (2022) Effect of Anti-Osteoporotic Treatments on Circulating and Bone MicroRNA Patterns in Osteopenic ZDF Rats International Journal of Molecular Sciences
- Fernandez GJ, Ramírez-Mejía JM, Urcuqui-Inchima S. (2022) Vitamin D boosts immune response of macrophages through a regulatory network of microRNAs and mRNAs. J. Nutr. Biochem.
- Gutmann, C., et al. (2022) Association of cardiometabolic microRNAs with COVID-19 severity and mortality. Cardiovascular Research
- Catalog Number
500-00012-SOM - Supplier
RealSeq Biosciences - Size
- Shipping
Dry Ice & Blue Ice

