Targeted Bisulfite Sequencing
Product Description
Targeted Bisulfite Sequencing is an NGS method used to evaluate site-specific DNA methylation changes. The process is similar to pyrosequencing but offers a much higher throughput overall. Our next-generation sequencing platforms can deliver a great amount of useful DNA methylation information with publication-ready data parsed by our expert bioinformatics scientists. The methylation analysis at single-base resolution of individual cytosine in DNA is facilitated by bisulfite treatment of DNA followed by PCR amplification of targeted region, library construction, and sequencing of the amplicon regions. Specific primers are designed for the region of interest and cytosine methylation changes are evaluated within that region. Each DNA methylation site of interest receives a high-sequencing depth of coverage for accurate, quantitative and single-base resolution data output.
EpiGentek’s Targeted Methyl-Seq platform yields reliable information on the methylation states of individual cytosines within the targeted region by effectively and efficiently preparing converted DNA for use in next-generation sequencing techniques.
Features
End-to-End Package
Includes everything from sample preparation to data analysis
Bioinformatics
FASTQ raw data, FASTQC quality control insights, read mapping, methylation calling and differential methylation analysis
Sequencing & Library Preparation Guarantee
Library QC by Bioanalyzer, KAPA quantification and Q30 sequencing score > 75%
High Sequencing Depth
Each base can be covered > 100X
Post-Completion Technical Support
Complimentary high-quality support to assist with data analysis at no additional charge
Please contact us to discuss your project and get a quote.
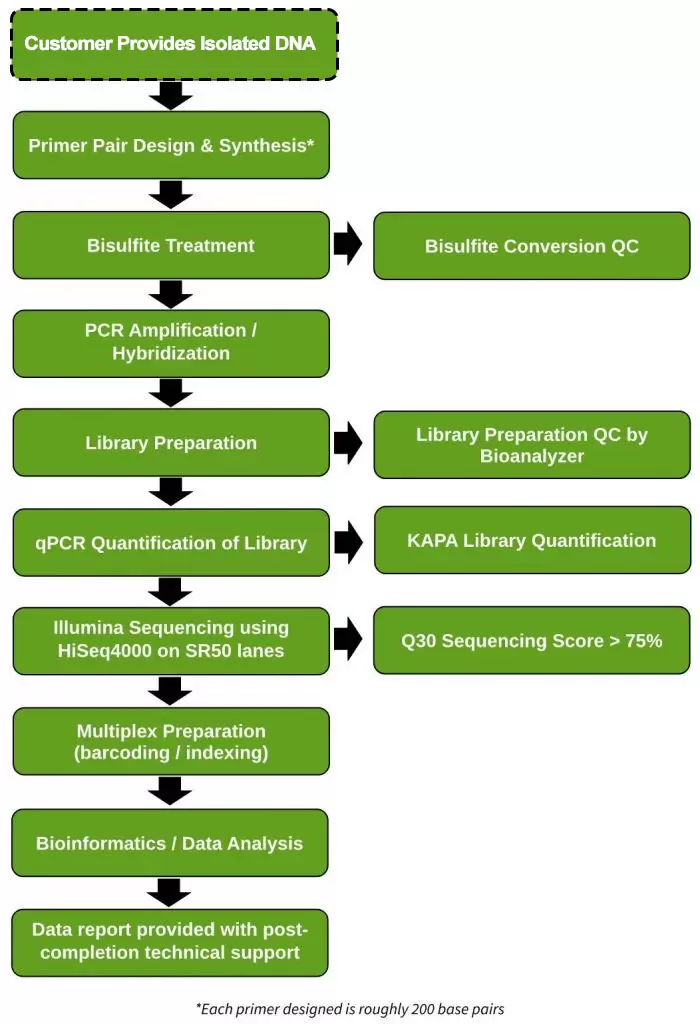
Workflow
Supporting Data
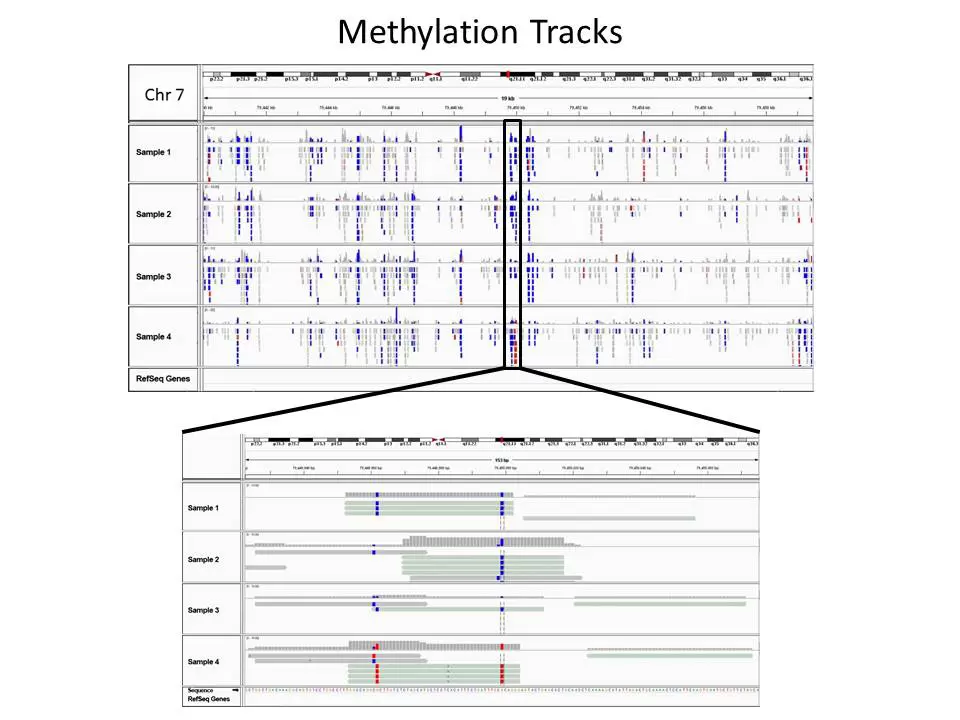
Fig.1. Read alignment views of DNA Methylation Analysis data at 19 kb and base pair resolution in chromosome 7 using Integrative Genomics Viewer (IGV). The data shows CpG methylation differences for treated and control samples. Control samples are unmethylated (blue) while treated samples are methylated (red).
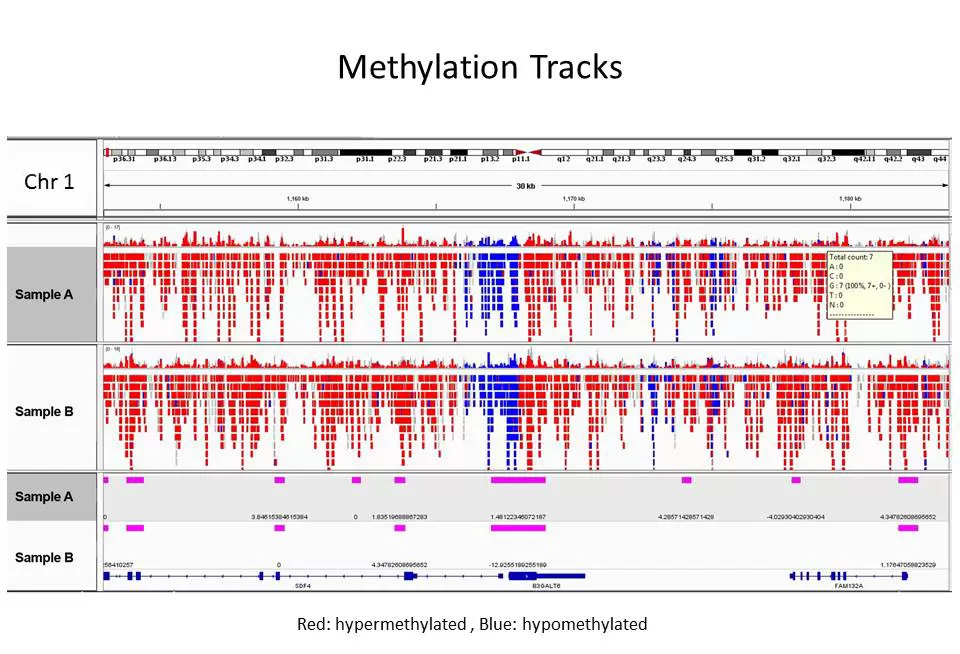
Fig.2. The Methylation Heatmap offers statistical grouping and visualization of samples based on DNA methylation levels of CpG sites. Green = 100% methylated; Red = 0% methylated.
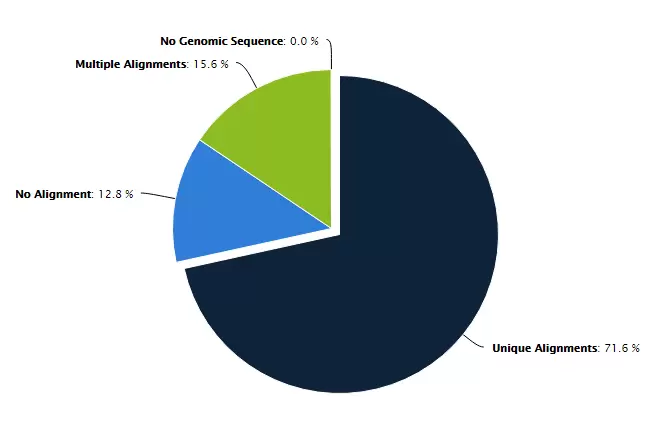
Fig.3. Bismark Mapping aligns bisulfite treated sequences to a genome of interest and will output comprehensive methylation data that can be visualized in a genome viewer or used for further analysis.
- Catalog Number
S-1TMS-EP - Supplier
EpigenTek - Size
- Shipping
RT
- Price
- Please inquire

