EpiQuik Histone H4 Modification Multiplex Assay Kit (Colorimetric)
Product Description
The EpiQuik™ Histone H4 Modification Multiplex Assay Kit (Colorimetric) is a complete set of optimized reagents to detect and quantify nearly all histone H4 modifications (10 different types) simultaneously in a simple, ELISA-like format with use of a standard microplate reader. The kit has the following advantages and features:
- Simultaneously screen and measure 10 different histone H4 modifications, which includes nearly all modified histone H4 sites.

- Quick and efficient procedure, which can be finished within 2 hours and 30 minutes.
- Innovative colorimetric assay without the need for radioactivity, electrophoresis, chromatography, or expensive equipment.
- High sensitivity with a detection limit as low as 0.5 ng/well for each modification pattern and a detection range from 20 ng to 500 ng/well of histone extracts.
- An assay control is conveniently included for quantification of each histone H4 modification.
- Total histone H4 sets are included, which can be used for normalizing total histone H4 levels for relative comparison of histone H4 content between different samples or different treatment conditions
- Strip-well microplate format makes the assay flexible: manual or high throughput, which enables analysis of a single modification or a total of 10 modification patterns within the same samples.
- Two extra 8-well strips are included in the kit which can be used, if necessary, for sample amount pre-optimization to determine the input amount (ex: 50, 100, 200 ng/well) needed to fall within the detection limits of the assay. Extra strips may also be used as assay controls and total histone level controls if selective detection of some histone H4 modifications from the total modification pattern is desired.
- Simple, reliable, and consistent assay conditions.
View assay kit demonstration video:
Background Information
Histone modifications have been defined as epigenetic modifiers. Post-translational modifications (PTMs) of histones include the acetylation of specific lysine residues by histone acetyltransferases (HATs), deacetylation by histone deacetylase (HDACs), the methylation of lysine and arginine residues by histone methytransferases (HMTs), the demethylation of lysine residues by histone demethylases (HDMTs), and the phosphorylation of specific serine groups by histone kinases (HKs). Additional histone modifications include the attachment of ubiquitin (Ub), small ubiquitin-like modifiers (SUMOs), and poly ADP-ribose (PAR) units. Next to DNA methylation, histone acetylation and histone methylation are the most well characterized epigenetic marks. There are many known sites and types of post-translational modification on human histone H4 that include Ser-1 phosphorylation, Arg-3 methylation, lysine 5, 8, 12, and 16 acetylation and lysine 20 mono-, di-, and trimethylation. Generally, euchromatin is characterized by a high level of histone H4 acetylation, which is mediated by histone acetyltransferases. Histone deacetylases have the ability to remove this epigenetic mark, which leads to transcriptional repression. Condensed heterochromatin is enriched in methylation of H4K20. Lysine residues of histone H4 can be mono-, di-, or trimethylated, each of which can differentially regulate chromatin structure and transcription. Along with other histone modifications such as phosphorylation, this enormous variation leads to a multiplicity of possible combinations of different modifications. This may constitute a “histone code”, which can be read and interpreted by different cellular factors.
Principle & Procedure
The EpiQuik™ Histone H4 Modification Multiplex Assay Kit (Colorimetric) is designed for measuring multiple histone H4 modifications simultaneously. In an assay with this kit, each histone H4 modified at specific sites will be captured by an antibody that is coated on the strip wells and specifically targets the appropriate histone H4 modification pattern. The captured histone modified at specific sites will be detected with a detection antibody, followed by a color development reagent. The ratio of modified histone is proportional to the intensity of absorbance measured by a microplate reader at a wavelength of 450 nm.
Starting Materials
Input materials can be histone extracts or purified histone H4 proteins obtained from human, mouse, rat, as well as a broad range of species including most plants, fungi, and bacteria, based on high sequence homology of histone H4. The amount of histone extracts for each assay can be 20 ng to 500 ng with an optimal range of 50 to 100 ng depending on the purity of histone extracts. The amount of purified histone H4 proteins for each assay can be 1 ng to 25 ng with an optimal range of 4 to 5 ng.
Performance Data
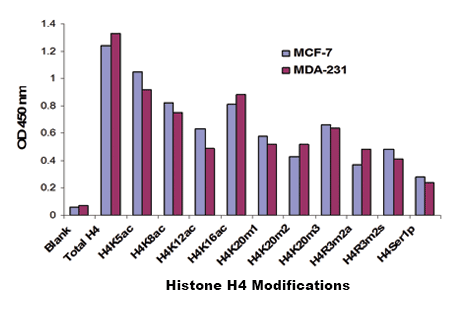
Fig. 2. Histone extracts were prepared from MCF-7 and MDA-231 cells using the EpiQuik™ Total Histone Extraction Kit (Cat. No. OP-0006) and 10 histone H4 modifications were measured using the EpiQuik™ Histone H4 Modification Multiplex Assay Kit (Colorimetric). 100 ng of total histone proteins per well were used.
Product Citations
- Cristian RE, Balta C, Herman H, Trica B, Sbarcea BG, Hermenean A, Dinischiotu A, Stan MS (2024) In Vivo Assessment of Hepatic and Kidney Toxicity Induced by Silicon Quantum Dots in Mice. Nanomaterials (Basel)
- Zhao Y, Wu X, Yang Y, Zhang L, Cai X, Chen S, Vera A, Ji J, Boström KI, Yao Y (2024) Inhibition of endothelial histone deacetylase 2 shifts endothelial-mesenchymal transitions in cerebral arteriovenous malformation models. J Clin Invest
- Olichwier A, Sowka A, Balatskyi VV, Gan AM, Dziewulska A, Dobrzyn P (2023) SCD1-related epigenetic modifications affect hormone-sensitive lipase (Lipe) gene expression in cardiomyocytes. Biochim Biophys Acta Mol Cell Res
- Krishnan R, Jang YS, Kim JO, Yoon SY, Rajendran R, Oh MJ (2022) Temperature dependent cellular, and epigenetic regulatory mechanisms underlying the antiviral immunity in sevenband grouper to nervous necrosis virus infection. Fish Shellfish Immunol
- Sugar SS, Heyob KM, Cheng X, Lee RJ, Rogers LK (2020) Perinatal inflammation alters histone 3 and histone 4 methylation patterns: Effects of MiR-29b supplementation. Redox Biol
- Catalog Number
P-3102-96-EP - Supplier
EpigenTek - Size
- Shipping
Blue Ice

