EpiNext ChIP-Seq High-Sensitivity Kit (Illumina)
The optimized protocol and components of the kit allow capture of low abundance protein/DNA complexes with minimized non-specific background levels and the ability to construct both non-barcoded (singleplexed) and barcoded (multiplexed) ChIP-Seq libraries quickly with reduced bias.
Product Description
The EpiNext™ ChIP-Seq High-Sensitivity Kit (Illumina) is a complete set of reagents required for carrying out a successful ChIP-Seq starting from mammalian cells or tissues.
The kit is designed to selectively enrich a chromatin fraction containing specific DNA sequences from various species, particularly mammals, and to prepare a ChIP-Seq library for next generation sequencing using Illumina platforms such as Illumina Genome Analyzer II, HiSeq and MiSeq systems.
The optimized protocol and components of the kit allow capture of low abundance protein/DNA complexes with minimized non-specific background levels and the ability to construct both non-barcoded (singleplexed) and barcoded (multiplexed) ChIP-Seq libraries quickly with reduced bias.
Features & Advantages
- Optimized buffers and protocol allow minimal ChIP background.
- Increased antibody selectivity and capture efficiency.
- Highly efficient enrichment of targeted DNA. Enrichment ratio of positive to negative control > 500.
- Usable for both non-barcoded (singleplexed) and barcoded (multiplexed) DNA library preparation.
- Input cell number range from 50,000 to 1,000,000 cells.
- Broad range of cell/tissue samples, including samples with limited or tiny amounts of available chromatin.
- Fast and streamlined procedure.
- Highly convenient - contains all required components for each step of ChIP-Seq.
- Minimized bias.
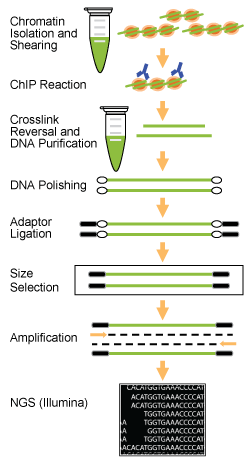
Fig. 1. Workflow of the EpiNext™ ChIP-Seq High-Sensitivity Kit (Illumina).
Background Information
Protein-DNA interaction plays a critical role for cellular functions such as signal transduction, gene transcription, chromosome segregation, DNA replication and recombination, and epigenetic silencing. Identifying the genetic targets of DNA binding proteins and knowing the mechanisms of protein-DNA interaction on a genome-wide scale is important for understanding cellular processes.
Chromatin immunoprecipitation (ChIP) followed by next generation sequencing (ChIP-Seq) offers an advantageous tool for studying genome-wide protein-DNA interactions. It allows for detection that a specific protein binds to specific sequences in living cells. In particular, ChIP antibodies targeted against various transcriptional factors (TF) for genome-wide transcription factor binding site analysis by Chip-Seq is in high demand.
Such analysis requires that ChIPed DNA contain minimal background for reliably identifying true TF-enriched regions. Currently used ChIP-Seq methods play an important role in identifying genome-wide protein-DNA interaction. However, these methods still have several drawbacks:
- (1) large amounts of cell/tissues are needed for obtaining a sufficient yield of library DNA, therefore these methods cannot be used for biological samples such as tumor biopsy and embryonic tissues whose amounts are limited;
- (2) the background levels of ChIPed DNA are high; and
- (3) the procedures are time consuming (>3 days) and inconvenient.
To address this issue, Epigentek developed the EpiNext ChIP-Seq High Sensitivity Kit (Illumina) by combining its microplate-based ultra ChIP and high sensitive DNA library construction technologies.
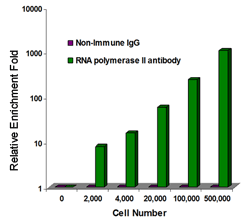
Fig. 2. Highly sensitive ChIP: The sheared chromatin isolated from different amounts of MBD-231 cells was used for ChIP-qPCR analysis of RNA polymerase II enrichment in GAPDH promoters.
Principle and Procedure
The EpiNext ChIP-Seq High-Sensitivity Kit (Illumina) contains all necessary reagents required for carrying out a successful ChIP-Seq starting from mammalian cells or tissues. In the ChIP reaction, chromatin is isolated from cell/tissues and the target protein-DNA complex is immunoprecipitated using the antibody of interest.
Immunoprecipitated DNA is then cleaned, released, and eluted. A positive control antibody (RNA polymerase II), a negative control non-immune IgG, and GAPDH primers are included in the kit, which can be used as a positive control to demonstrate the efficacy of the kit reagents and protocol. RNA polymerase II is considered to be enriched in the GAPDH gene promoter that is expected to be undergoing transcription in most growing mammalian cells and can be immunoprecipitated by RNA polymerase II antibody but not by non-immune IgG. In the library preparation, ChIPed DNA fragments are end repaired and dA tailed (end polishing) simultaneously.
Adaptors are then ligated to both ends of the polished DNA fragments for amplification and sequencing. Ligated fragments are size selected and purified using MQ binding beads, which allows quick and precise size selection of DNA. Size-selected DNA fragments are amplified with a high-fidelity PCR mix which ensures maximum yields from minimum amounts of starting material and provides highly accurate amplification of library DNA with low error rates and minimum bias.
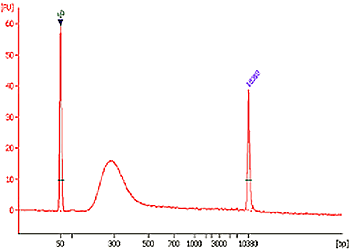
Fig. 3. Size distribution of library fragments. Ten nanograms of DNA was ChIPed by RNA polymerase
Starting Materials
Starting materials can include various tissue or cell samples such as culture cells from a flask or plate, fresh and frozen tissues, etc.
Product Citations
- Cheng, V. et. al. (2021) Utilizing systems biology to reveal cellular responses to peroxisome proliferator-activated receptor γ ligand exposure Current Research in Toxicology
- Upadhyaya B et al. (2017) Prenatal Exposure to a Maternal High-Fat Diet Affects Histone Modification of Cardiometabolic Genes in Newborn Rats. Nutrients 9(4)

