EpiNext CUT&RUN Fast Kit
Product Description
- Quick and reliable recovery of target protein-enriched regions
- Input cell amount as few as 500 cells, or chromatin amount as low as 0.1 µg
- Minimized immunocapture background for sequencing data analysis with <10 million reads
- Fast, streamlined procedure in less than 3h
- Highly convenient - all required components included
Overview
There are several advanced methods available for ChIP-Seq with reduced cell numbers or increased resolution. These methods include ChIP-exo and ChIPmentation. ChIP-exo provides high-resolution mapping, but is time-consuming and requires an ample supply of input cells. While ChIPmentation uses transposase and sequencing-compatible adaptors to enable the integration of ligation during the ChIP process, it follows a traditionally slow (2 days) ChIP procedure and cannot achieve high-resolution mapping. CUT&RUN (Cleavage Under Target & Release Using Nuclease) was developed to release the captured target protein/DNA complex from limited biological materials for mapping protein-DNA interaction, and it has significantly improved mapping resolution. However, CUT&RUN needs expensive PA/Mnase fusion protein that has significant A/T sequence bias, causing the target protein-interacted DNA region profiles to be seriously affected by the level of MNase digestion. Therefore, as an improvement, we have developed a new technique: cleavage under target and recover using nuclease fast (CUT&RUN Fast) for enriching histone or TF-bound DNA. Our innovative approach combines the advantages of ChIP-exo and CUT&RUN with the fastest procedures and incorporates it into the EpiNext™ CUT&RUN Fast Kit.
Principle & Procedure
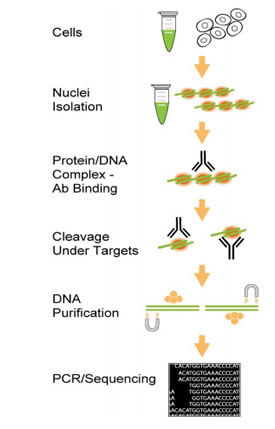
The kit contains all necessary reagents required for carrying out a successful CUT&RUN starting from mammalian cells or isolated nuclei/chromatin. In the reaction, nuclei are isolated from cells. The target protein-DNA complex is bound/captured with the CHIP-grade antibody of interest. With the use of a unique nucleic acid cleavage enzyme mix, chromatin is fragmented, and DNA sequences in both ends of the target protein/DNA complex are cleaved/removed. At the same time, the DNA sequence occupied by the target protein is unaffected. The target protein-bound DNA is then purified and eluted. The enriched DNA can be used for the analysis of protein-DNA interaction with various methods, especially with next generation sequencing. Included in the kit are a positive control antibody (anti-H3K9me3), a negative control non-immune IgG, and control chromatin, which can be used to demonstrate the efficacy of the kit and performance at the PCR/bioanalyzer step.
Performance Data

Fig 2. Target protein/DNA enrichment using the EpiNext CUT&RUN Fast Kit:
Histone/DNA complex was captured by the control antibodies (anti-H3K9me3 and anti-RNA polymerase II, Epigentek) from 100,000 Jurkat cells. Non-immuno IgG was used as control. The enriched DNA was purified and fluorescently quantified for enrichment fold comparison.
Product Citations
- Fritzke M, Chen E, Chen K, Tang W, Stinson S, Ito M, Xu L, (2023) The MYC-YBX1 Circuit in Maintaining Stem-like Vincristine-Resistant Cells in Rhabdomyosarcoma 0
- McFadden T, Gaito N, Carucci I, Fletchall E, Farrell K, Jarome TJ (2023) Controlling hypothalamic DNA methylation at the Pomc promoter does not regulate weight gain during the development of obesity. PLoS One
- Tao Y, Zhao J, Yin J, Zhou Z, Li H, Zang J, Wang T, Wang Y, Guo C, Zhu F, Dai S, Wang F, Zhao H, Mao H, Liu F, Zhang L, Wang Q (2023) Hepatocyte TIPE2 is a fasting-induced Raf-1 inactivator that drives hepatic gluconeogenesis to maintain glucose homeostasis. Metabolism
- Catalog Number
P-2028-24-EP - Supplier
EpigenTek - Size
- Shipping
Blue Ice

