MethylFlash Hydroxymethylated DNA (5-hmC) Quantification Kit (Colorimetric)
Product Description
- Rapid procedure, completed within 3 hours and 45 minutes
- Colorimetric assay with easy-to-follow steps for convenience and speed
- High sensitivity - detection limit as low as 20 pg of hydroxymethylated DNA
- High specificity - no cross-reactivity to methylcytosine and unmethylated cytosine, 5-hydroxymethylcytosine is separately detected
- Universal positive and negative controls included
- Flexible strip-well microplate format
- Simple, reliable, and consistent assay conditions
The MethylFlash™ Hydroxymethylated DNA Quantification Kit is a complete set of optimized buffers and reagents to quantify global DNA hydroxymethylation by specifically measuring levels of 5-hydroxymethylcytosine (5-hmC) without cross-reactivity to methylcytosine and unmethylated cytosine.
5-hmC is a modified form of 5-cytosine, recently discovered in animal tissues. The function of 5-hmC in epigenetics may be different from its forerunner 5-methylcytosine (5-mC) and currently remains a mystery. It is believed, though, that 5-hmC plays an important role in switching genes on and off. The presence of 5-hmC makes it necessary to not only re-evaluate existing DNA methylation data, but also necessary to determine relative distribution and changes of 5-hmC in human tissues of healthy and diseased statuses. Prior to Epigentek’s MethylFlash™ technology, there were no methods that could be used for practically and routinely identifying 5-hmC and discriminating this base from 5-mC.
Distinguishing between 5-hmC and 5-mC
Currently used methylated DNA analysis methods including restriction enzyme digestion and bisulfite or MeDIP-mediated MS-PCR and sequencing are not suitable for 5-hmC detection as 5-hmC and 5-mC are virtually indistinguishable with these methods. To address this problem, the MethylFlash™ Hydroxymethylated DNA Quantification Kit uses a unique procedure to quantify global DNA hydroxymethylation. This product provides a cost-effective way to measure levels of 5-hydroxymethylcytosine and to distinguish between 5-hmC, 5-mC, and C. This allows for researchers to re-evaluate their DNA methylation data for DNA hydroxymethylation and to efficiently look for DNA hydroxymethylation in new DNA samples.
Assay Procedure

High Sensitivity

Synthetic hydroxymethylated DNA was added into the assay wells at different concentrations and then measured with the MethylFlash™ Hydroxymethylated DNA Quantification Kit. The detection limit of the input DNA can be as low as 20 pg of hydroxymethylated DNA with a dynamic range from 40 pg to 20 ng. Analytic sensitivity is about ten-fold higher than that obtained by HPLC (0.008% vs 0.08%).
High Specificity
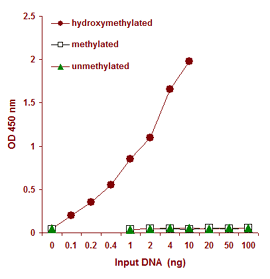
Synthetic unmethylated DNA (contains only 5-cytosine), methylated DNA (contains only 5-methylcytosine), and hydroxymethylated DNA standard (contains only 5-hydroxymethylcytosine) were added into the assay wells at different concentrations and then measured with the MethylFlash™ Hydroxymethylated DNA Quantification Kit. There is no cross-reactivity to methylcytosine and unmethylated cytosine so that only hydroxymethylated DNA (5-hmC) is detected.
Product Citations
- Ganguly A, Ghosh S, Shin BC, Touma M, Wadehra M, Devaskar SU (2024) Gestational exposure to air pollutants perturbs metabolic and placenta-fetal phenotype. Reprod Toxicol
- Sint Jago SC, Bahabry R, Schreiber AM, Homola J, Ngyuen T, Meijia F, Allendorfer JB, Lubin FD (2023) Aerobic exercise alters DNA hydroxymethylation levels in an experimental rodent model of temporal lobe epilepsy. Epilepsy Behav Rep
- Stoccoro A, Nicolì V, Coppedè F, Grossi E, Fedrizzi G, Menotta S, Lorenzoni F, Caretto M, Carmignani A, Pistolesi S, Burgio E, Fanos V, Migliore L (2023) Prenatal Environmental Stressors and DNA Methylation Levels in Placenta and Peripheral Tissues of Mothers and Neonates Evaluated by Applying Artificial Neural Networks. Genes (Basel)
- Arora I, Li S, Crowley MR, Li Y, Tollefsbol TO (2022) Genome-Wide Analysis on Transcriptome and Methylome in Prevention of Mammary Tumor Induced by Early Life Combined Botanicals. Cells
- Dar TU, Akhter N, Dar SA (2022) Editorial: Epigenomic polymorphisms: The drivers of diversity and heterogeneity. Front Genet
- Catalog Number
P-1036-96-EP - Supplier
EpigenTek - Size
- Shipping
Blue Ice

