MethylFlash Global DNA Hydroxymethylation (5-hmC) ELISA Easy Kit (Colorimetric)
Product Description
The MethylFlash™ Global DNA Hydroxymethylation (5-hmC) ELISA Easy Kit (Colorimetric) is a complete set of optimized buffers and reagents to colorimetrically quantify global DNA hydroxymethylation status by specifically measuring levels of 5-hydroxymethylcytosine (5-hmC) in a simplified, “one-step" ELISA-like format. As a fourth generation technology of Epigentek's popular global DNA methylation technique, it is a further refinement of the predecessor MethylFlash kit by improving upon speed, simplicity, sensitivity, and reproducibility.
- Fast - Reduced steps so that the entire procedure only needs 2 hours*
- Robust - Improved kit composition allows the assay to have a greater “signal window" with reduced variation between replicates
- Convenient - Inherently low background noise, thereby eliminating the need for DNA denaturation and plate blocking steps
- Sensitive - Detection limit can be as low as 0.01% hydroxymethylated DNA from 100 ng of input DNA
- Specific - High specificity to 5-hmC, with no cross-reactivity to unmethylated cytosine or methylated cytosine within the indicated concentration range of the sample DNA
- Universal – Positive and negative controls and allow detection of DNA hydroxymethylation in any species from either single-stranded or double-stranded input DNA
- Accurate - Optimized positive controls that can be fractionalized in percentage scale, allowing the assay to be more accurate and highly comparable with HPLC-MS analysis
- Flexible - Strip-well microplate format makes the assay available for manual or high throughput analysis
* Based on a single sample assay in duplicate
Background
5-hydroxymethylcytosine (5-hmC), as a sixth DNA base with functions in transcription regulation, has been detected to be abundant in human and mouse brains and embryonic stem (ES) cells. In mammals, it can be generated by the oxidation of 5-mC, a reaction mediated by the ten-eleven translocation (TET) family of 5-mC-hydroxylases. 5-hmC has been demonstrated to be tissue specific, ranging from undetectable levels in cultured cell lines to 0.6% in human brain tissues, and can be as high as 8% of total DNA in some other species. The biological significance of 5-hmC as an important epigenetic modification in phenotype and gene expression has been recently recognized. For example, global decrease in 5-hmC content (DNA hypomethylation) has been exhibited in nearly all cancers and has been proposed as a molecular marker and therapeutic target in cancer as well.
Principle & Procedure
This kit contains all reagents necessary for the quantification of global DNA hydroxymethylation. In this assay, DNA is bound to strip-wells that are specifically treated to have a high DNA affinity. The hydroxymethylated fraction of DNA is detected using a 5-hmC mAb-based detection complex in a one-step manner and then quantified colorimetrically by reading the absorbance in a microplate spectrophotometer. The percentage of hydroxymethylated DNA is proportional to the OD intensity measured.
Starting Materials
Input DNA should be relatively pure with 260/280 ratio >1.6 and can be diluted with water or TE buffer. The DNA amount can range from 20 ng to 200 ng per reaction. However, we recommend using 100 ng of DNA, which is the optimized input amount for the best results.
Performance Data
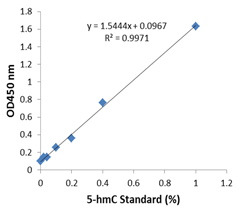
Fig.2. Example of an optimal standard curve generated with 5-hmC standard control.
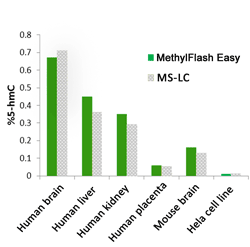
Fig.3. Accurate quantification of 5-hmC content of various DNA samples from different species with the MethylFlash™ Global DNA Hydroxymethylation (5-hmC) ELISA Easy Kit (Colorimetric). The results are closely correlated with those obtained by MS-LC.
Product Citations
- Angelin M, Gopinath P, Raghavan V, Thara R, Ahmad F, Munirajan AK, Sudesh R (2024) Global DNA and RNA Methylation Signature in Response to Antipsychotic Treatment in First-Episode Schizophrenia Patients. Neuropsychiatr Dis Treat
- Jiang X, Xia L, Tang T, Fan X, Wang R, Wang M, Yang W, Yan J, Qi K, Li P (2024) Decreased vitamin D bio-availability with altered DNA methylation of its metabolism genes in association with the metabolic disorders among the school-aged children with degree I, II and III obesity. J Nutr Biochem
- Lu X, Mao J, Qian C, Lei H, Mu F, Sun H, Yan S, Fang Z, Lu J, Xu Q, Dong J, Su D, Wang J, Jin N, Chen S, Wang X (2024) High estrogen during ovarian stimulation induced loss of maternal imprinted methylation that is essential for placental development via overexpression of TET2 in mouse oocytes. Cell Commun Signal
- Thomas D, Palczewski M, Kuschman H, Hoffman B, Yang H, Glynn S, Wilson D, Kool E, Montfort W, Chang J, Petenkaya A, Chronis C, Cundari T, Sappa S, Islam K, McVicar D, Fan Y, Chen Q, Meerzaman D, Sierk M (2024) Nitric oxide inhibits ten-eleven translocation DNA demethylases to regulate 5mC and 5hmC across the genome. Res Sq
- Wei X, Li J, Cheng Z, Wei S, Yu G, Olsen ML (2024) Decoding the Epigenetic Landscape: Insights into 5mC and 5hmC Patterns in Mouse Cortical Cell Types. bioRxiv
- Catalog Number
P-1032-48-EP - Supplier
EpigenTek - Size
- Shipping
Blue Ice

