CRISPRclean Plus Stranded Total RNA Prep with rRNA Depletion
Identify lower expressing transcripts with CRISPRclean Plus
Product Description
Complex samples may have a mixture of bacterial and human cells, all actively expressing ribosomal RNA. Take advantage of the power of CRISPRclean for effective ribosomal RNA removal. CRISPRclean can also remove uninformative bacterial reads in-vitro ahead of sequencing – redistributing >80% of sequencing reads to enable identification of lower expressing, biologically relevant transcripts.
- Streamlined CRISPRclean workflow: stranded total RNA library prep with ribodepletion
- Ideal for analysis of samples containing complex mixtures of eukaryotic and bacterial rRNA
- 3 to 7-fold increase in coverage of SARS-CoV-2 and other genomes in nasopharyngeal (NSP) samples
- Single workflow to detect viral genomic data, microbiome composition, co-infections, and host gene expression
How it works
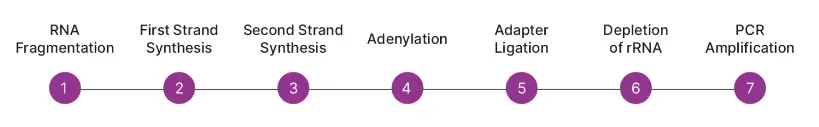
The workflow begins with RNA fragmentation through high-temperature incubation and continues to first and second strand synthesis, converting RNA fragments into cDNA libraries. The assay achieves 98% strand specificity by incorporating dUTP during second strand synthesis. Adenylation modifies the 3’ ends of the double-stranded cDNA and dATP prepares the library for adapter ligation. After ligating the unique dual index adapters, the library is ready for depletion. CRISPR depletion happens in two successive incubations. The first cleaves bacterial rRNAs and the second cleaves eukaryotic rRNAs. Cas9 and guide RNAs are combined to form the ribonucleoprotein complex that cuts the targeted rRNA sequences.
Cleaved rRNA cannot be amplified and is removed through size selection with magnetic beads.
Specifications
Related Product: CRISPRclean Unique Dual Index Adapter Plate for RNA Prep (Set A)
Performance Data
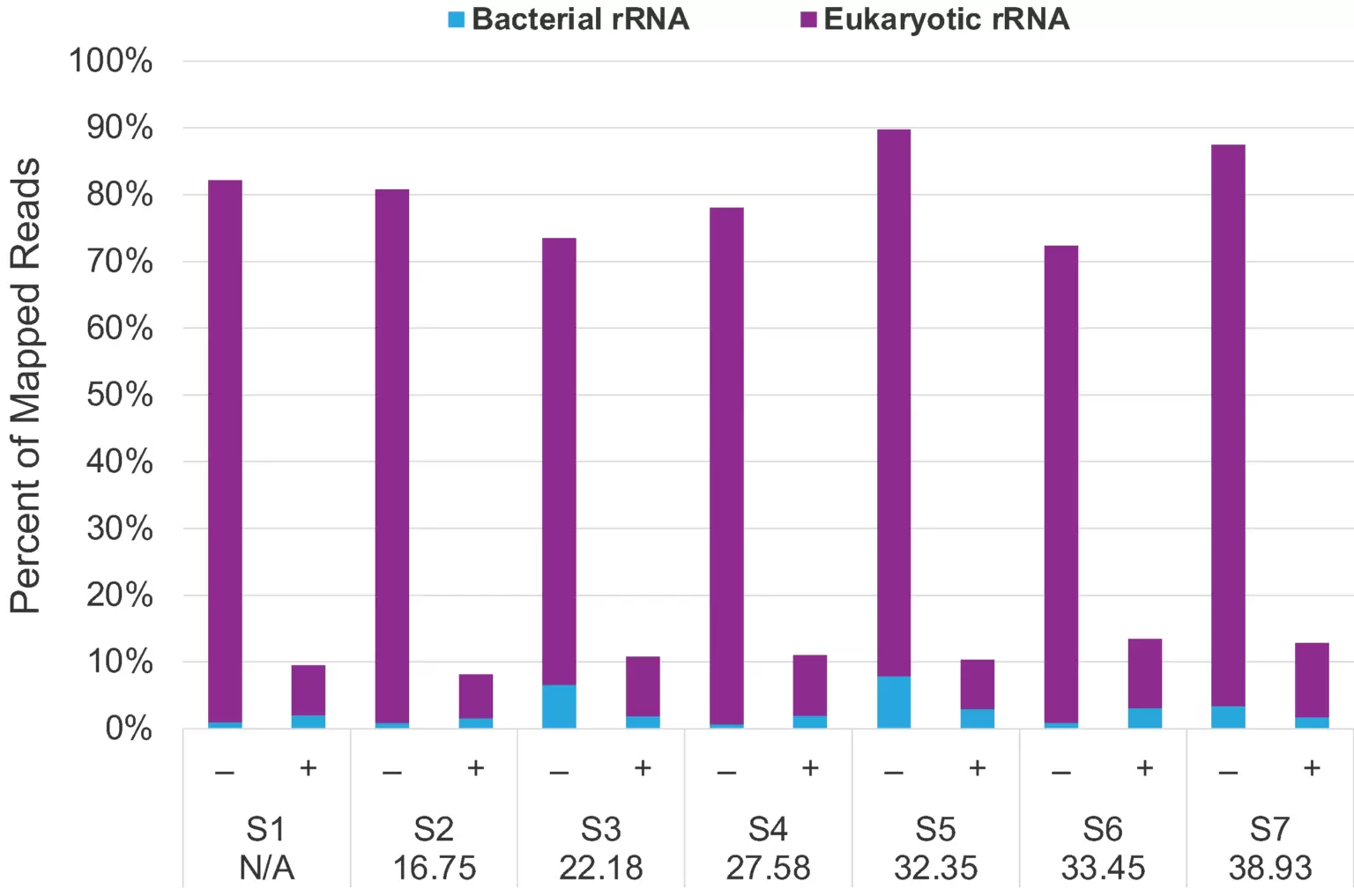
Fig.1. Before and after depletion comparison showed efficient >80% rRNA depletion of eukaryotic and bacterial using clinical NSP samples across different Ct values from negative to 38.93, which has very low viral load.
- Catalog Number
KIT1016-JCG - Supplier
Jumpcode Genomics - Size
- Shipping
Dry Ice

