Arima-HiC+ Kit
Accurate and Reproducible Genome Conformation Analyses (Epigenetics)
Product Description
The Arima-HiC approach uses a highly simplified and robust protocol that streamlines Hi-C to be a 6 hour workflow followed by library prep and and next-generation sequencing. Arima-HiC empowers researchers to leverage Hi-C technology to perform genome-wide and targeted indentification of promotor-enhancer interactions, link non-coding disease-associated variants to target promotors, and uncover how pertubations in genome conformation impact gene regulation in diseases contexts.
- Proven Performance - greater accuracy of chromatin loop and topological domain detection even at reduced sequencing depths
- Fast and user-friendly workflow - rapid 6 hour HiC-workflow
- Assured quality - rapid and easy library QC steps that accurately predict library quality, ensuring sequencing success
- Flexible input requirements - support for a broad range of sample types and an optimized protocol for low sample input
Arima-HiC also empowers researchers to leverage HiC-technology to detect and correct misjoin errors in draft assemblies, assign contigs to chromosomes de novo and order and orient contigs into chromosome scale scaffolds. It has been successfully performed on a wide-range of species from the plant and animal kingdoms.
Arima-HiC Workflow Overview
Chromatin from a sample source (tissues, cell lines, or blood) is first crosslinked to preserve the genome sequence and structure. The crosslinked chromatin is digested using a restriction enzyme (RE) cocktail. The 5’-overhangs are then filled in, causing the digested ends to be labeled with a biotinylated nucleotide. Next, spatially proximal digested ends of DNA are ligated, capturing the sequence and structure of the genome. The ligated DNA is then purified, producing pure proximally-ligated DNA. The proximally-ligated DNA is fragmented, and the biotinylated fragments are enriched. The enriched fragments are subjected to a custom library preparation protocol utilizing a range of supported commercially available library prep kits. Depending on the choice of library prep kit, a separate Arima-HiC Library Prep user guide is provided that contains a custom protocol for converting proximally-ligated DNA to Arima-HiC libraries.
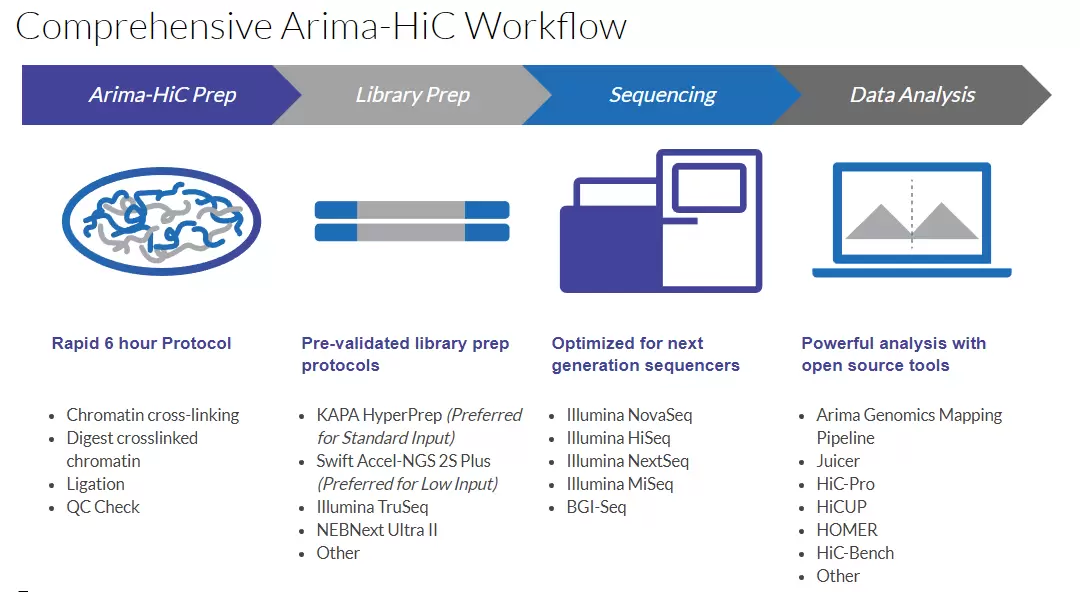
Sequencing and Data Analysis
Arima-HiC libraries are sequenced via Illumina® sequencers in “paired-end” mode. The tools necessary for analyzing the resulting data depend on the application.
Applications
- Identification and visualization of TADs, loops, or other features (see data sheet)
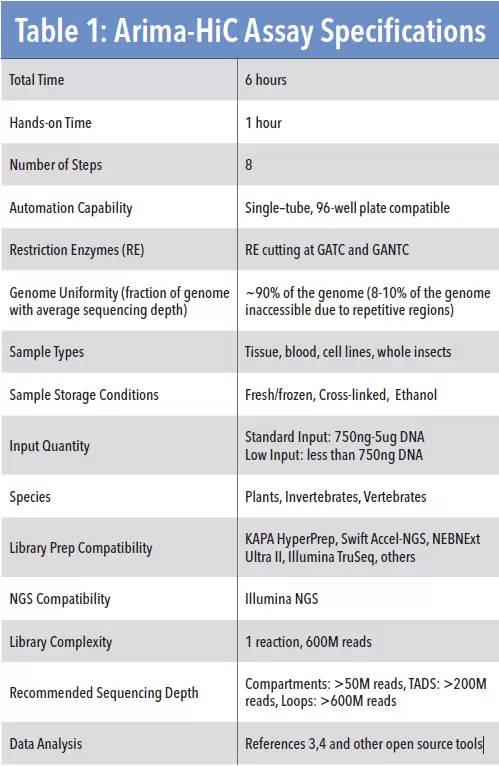
Specifications
Product Citations
- Shilpa Garg et al. (2020) Accurate chromosome-scale haplotype-resolved assembly of human genomes. bioRxiv 810341
- Arang Rhie, Shane A. McCarthy, Olivier Fedrigo, et al. (2020) Towards complete and error-free genome assemblies of all vertebrate species. bioRxiv 110833
- Baudry, L., Guiglielmoni, N., Marie-Nelly, H. et al (2020) instaGRAAL: chromosome-level quality scaffolding of genomes using a proximity ligation-based scaffolder. Genome Biol 21:148
- Chenfu Shi, Helen Ray-Jones, James Ding, Kate Duffus, et al. (2020) An active chromatin interactome in relevant cell lines elucidates biological mechanisms at genetic risk loci for dermatological traits. bioRxiv 973271
- Coline Arnould, Vincent Rocher, Thomas Clouaire, et al. (2020) Loop extrusion as a mechanism for DNA Double-Strand Breaks repair foci formation. bioRxiv 945311
- Catalog Number
A51008-ARI - Supplier
Arima - Size
- Shipping
Blue Ice

