LongPlex™ Long Fragment Multiplexing Kit, Any Index Set
Realize the potential of your PacBio long read sequencer
Product Description
Realize the throughput and cost-effectiveness of PacBio HiFi sequencing using seqWell’s LongPlex Long Fragment Multiplexing Kit. Its speed, simplicity, and scalability enable massive sample multiplexing that unleashes scalable long read sequencing.
- Plate-based, enzymatic method to simultaneously fragment & index genomic DNA for scalable PacBio HiFi sequencing
- Massive multiplexing using 96 unique dual indexes (UDI) that can be combinatorially expanded with PacBio indexing
- Early sample pooling greatly reduces all-in cost per sample for long read sequencing without sacrificing data quality
- Run more long read samples per flow cell without compromising PacBio HiFi sequencing quality
- Supports long read applications including microbial and small genome sequencing, metagenomics, low pass sequencing, and targeted hybrid capture
Transposase-based gDNA fragmentation ensures a consistent, scalable generation of 8 -10 kB fragment lengths. The enzymatic fragmentation using adapter-loaded transposase allows for simultaneous fragmentation and indexing of gDNA. Importantly, this eliminates the need for mechanical shearing of gDNA and greatly increases sample throughput.
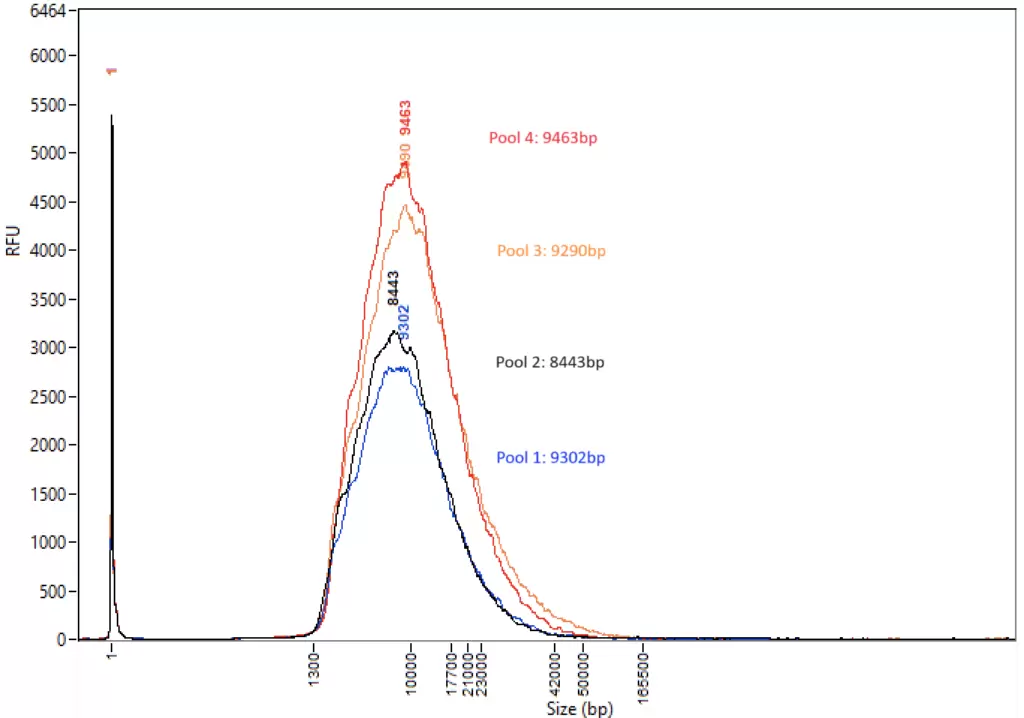
Figure 1: DNA samples were analyzed on the Femto Pulse system following LongPlex processing using the standard PCR-free protocol and SMRTbell 3.0 library preparation. The traces show reproducible fragmentation profiles across four 24-plexes (full plate of 96 samples pooled in 4 separate pools of 24) with an average fragment length >8 kb.
Kit Content
- Tagging Reagent Plate (96-well microtiter plate)
- 3X Coding Buffer
- X Solution
- Library Primary Mix for optional PCR
Simple Workflow for Scalable Fragmentation and Multiplexing
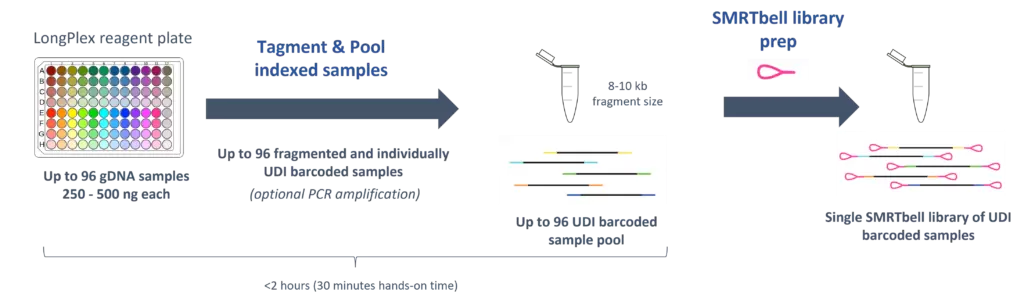
- Simultaneously fragment and index up to 96 gDNA samples with no DNA shearing required
- Plate-based protocol performed in < 2 hours with only 30 minutes of hands-on time
- Produces fragments ranging from 8 – 10 kb long
- Sample multiplexing via pooling of up to 96 samples prior to SMRTbell library preparation
- PCR-free and PCR-based protocols available to support a variety of needs including difficult samples
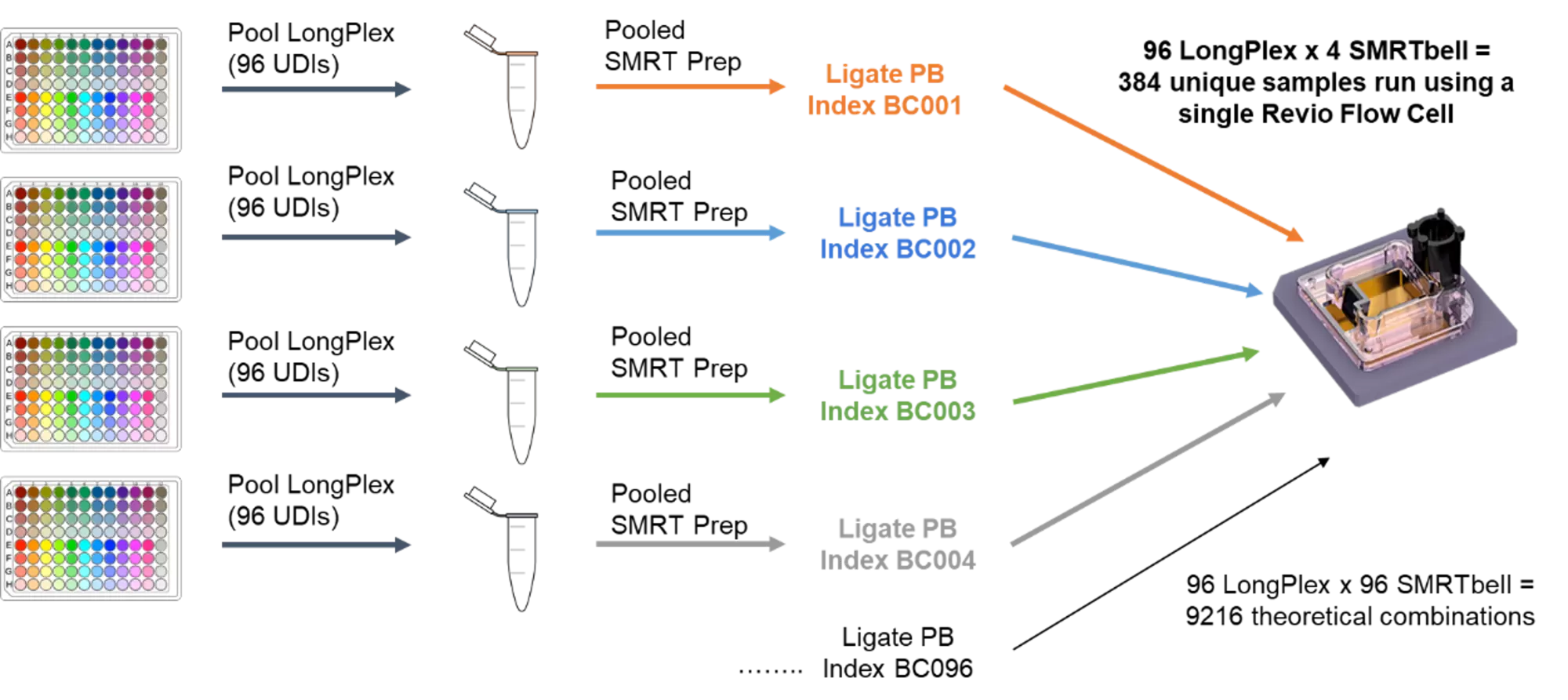
Reducing Long Read Sequencing Costs Through Massive Multiplexing
Efficiently multiplex more samples per flow cell without compromising sequencing quality on PacBio HiFi long-read sequencers. To run 384 long read samples, you no longer need to perform 384 PacBio SMRTbell library preparations! Simply use the plate-based LongPlex method, followed by 4 SMRTbell library preps enabling 384 sample on a single flow cell. Pooling samples early allows you to maximize sequencer throughput while driving down costs.
Specifications
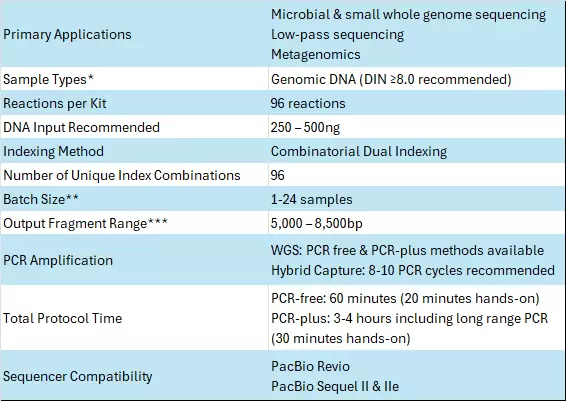
*Other sample types may be compatible. Please contact BioCat for guidance.
**Up to 96 samples can be pooled past LongPlex barcoding prior to PacBio SMRTbell but it may impact read output and coverage uniformity
***Fragment size will depend on magnetic bead cleanup ratios used
Additional Resources
Scalable Long-Read Low-Coverage Human Whole Genome Sequencing (lcWGS) for Genotype Imputation (poster)
An Efficient Enzyme-Based Library Workflow for Multiplexed Long Read Whole Genome Sequencing (poster)
- Catalog Number
301315-SQW - Supplier
seqWell - Size
- Shipping
Dry Ice

