H3K27ac Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag
Product Description
This H3K27ac (histone H3 acetylated at lysine 27) antibody meets EpiCypher’s lot-specific SNAP-Certified™ criteria for specificity and efficient target enrichment in both CUT&RUN and CUT&Tag applications. This requires <20% cross-reactivity to related histone PTMs determined using the SNAP-CUTANA™ K-AcylStat Panel of spike-in controls. High target efficiency is confirmed by consistent genomic enrichment at varying cell inputs: 500k and 50k cells in CUT&RUN; 100k and 10k cells in CUT&Tag. High efficiency antibodies display similar peak structures and highly conserved genome-wide signal even at reduced cell numbers. H3K27ac is associated with gene activation and is enriched at active enhancers and promoters [1].
Validation Data - CUT & Tag
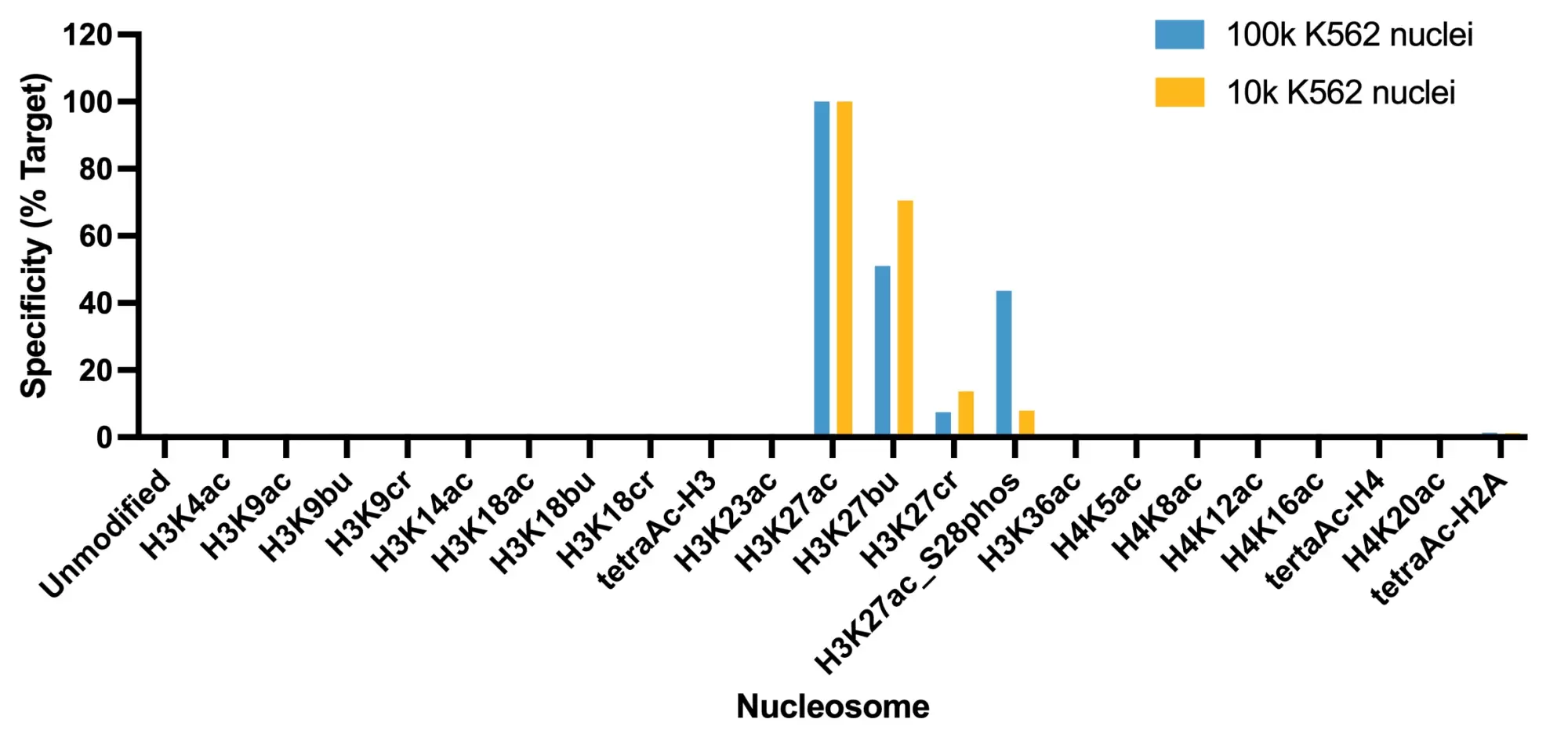
Figure 1: SNAP specificity analysis in CUT&Tag. CUT&Tag was performed and CUT&Tag sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the K-AcylStat panel (x-axis). Data are expressed as a percent relative to on-target recovery (H3K27ac set to 100%). The antibody showed recovery of H3K27ac spike-in nucleosomes and to a lesser extent H3K27ac nucleosomes that contain a proximal phosphorylation at S28 at both 500k and 50k cells. The antibody cross-reacts with butyrylation at H3K27, but this is typically low abundance in cells [2].
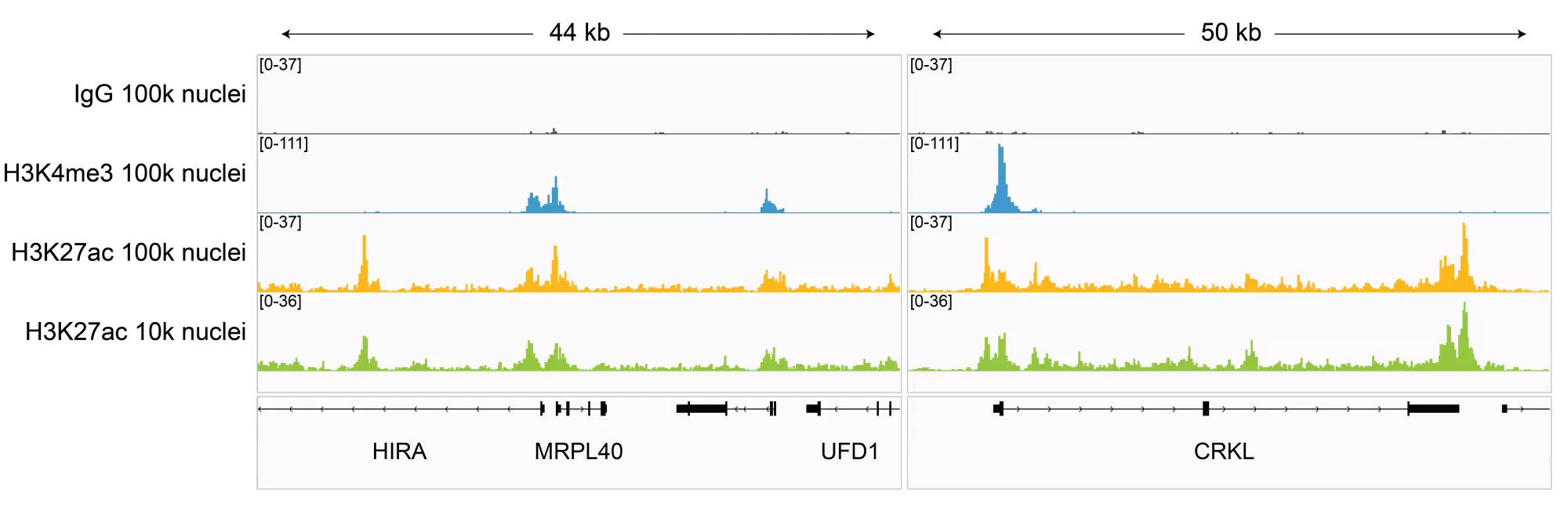
Figure 2: H3K27ac CUT&Tag representative browser tracks. CUT&Tag was performed and gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). H3K27ac antibody tracks display peaks at promoters and enhancers, consistent with the biological function of this PTM. Similar results in peak structure and location were observed for both 100k and 10k nuclei inputs.
Validation Data - CUT & RUN
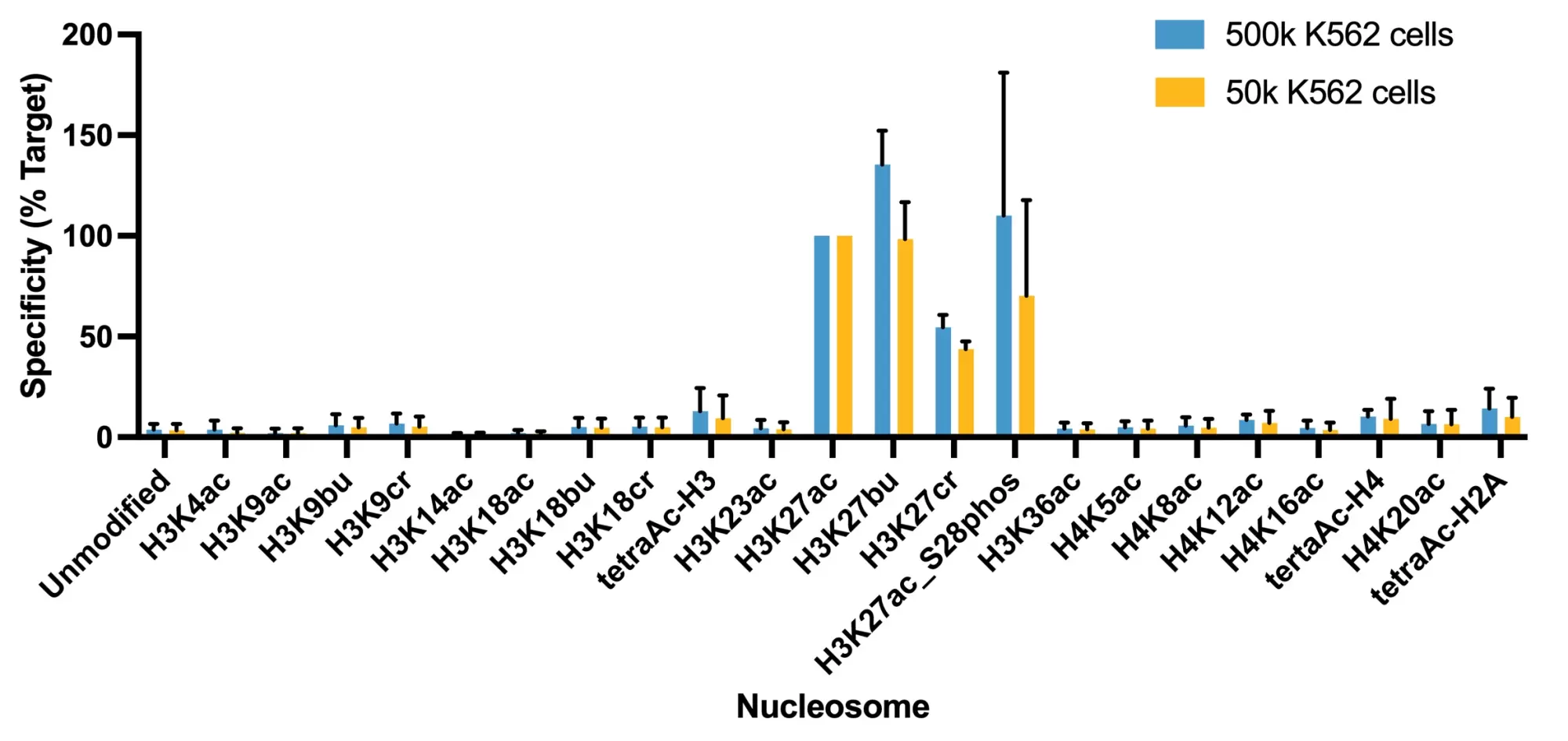
Figure 3: Average SNAP specificity analysis from two CUT&RUN experiments. CUT&RUN was performed and CUT&RUN sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the K-AcylStat panel (x-axis). Data are expressed as a percent relative to on-target recovery (H3K27ac set to 100%). The antibody showed recovery of H3K27ac spike-in nucleosomes as well as H3K27ac nucleosomes that contain a proximal phosphorylation at S28 at both 500k and 50k cells. The antibody cross-reacts with extended acyl states (butyrylation and crotonylation) at H3K27, but these are typically low abundance in cells [2].
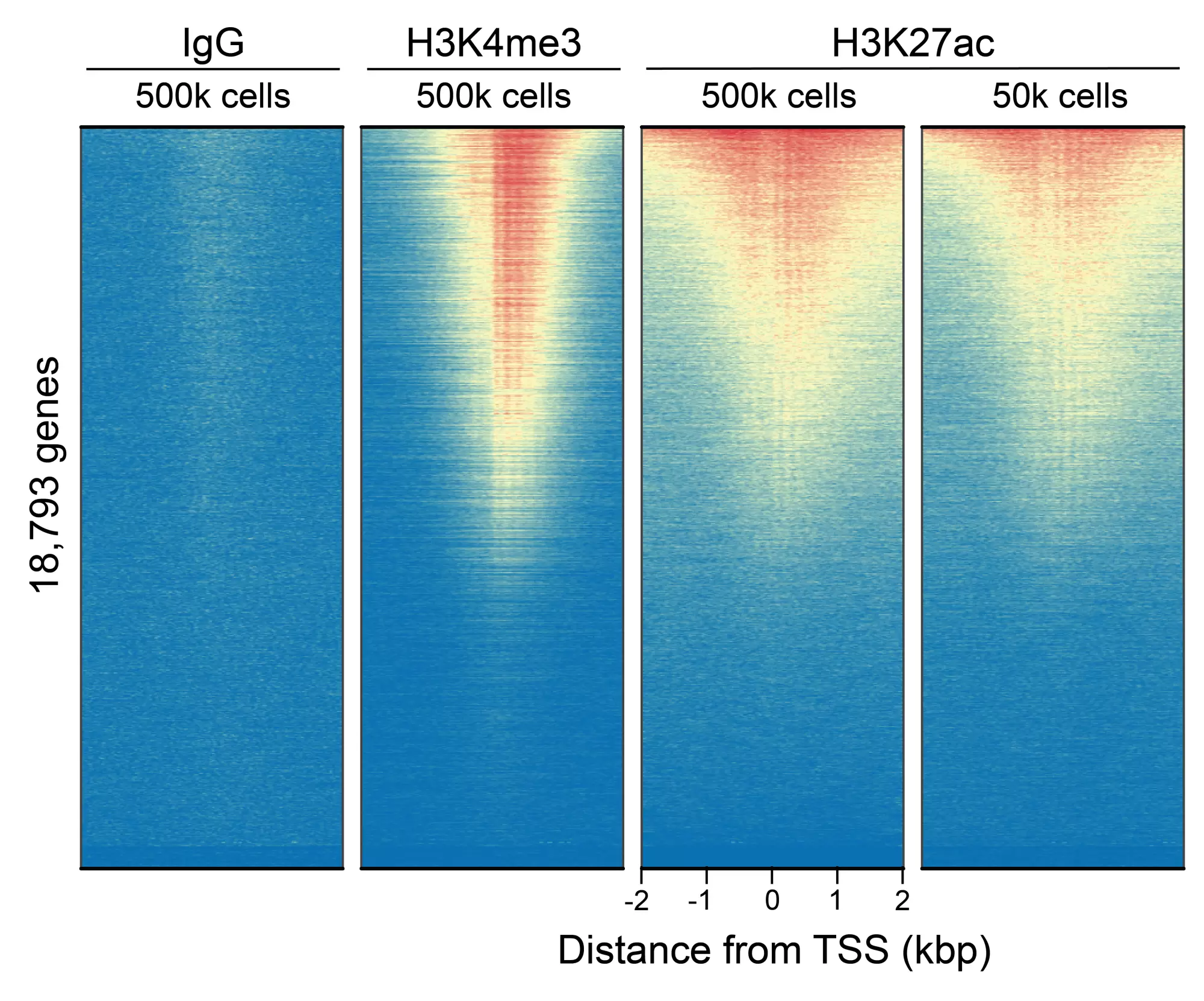
Figure 4: CUT&RUN genome-wide enrichment. CUT&RUN was performed and sequence reads were aligned to 18,793 annotated transcription start sites (TSSs ± 2 kbp). Signal enrichment was sorted from highest to lowest (top to bottom) relative to the H3K27ac - 500k cells reaction (all gene rows aligned). High, medium, and low intensity are shown in red, yellow, and blue, respectively. H3K4me3 positive control and H3K27ac antibodies produced the expected enrichment pattern, which was consistent between 500k and 50k cells and greater than the IgG negative control.
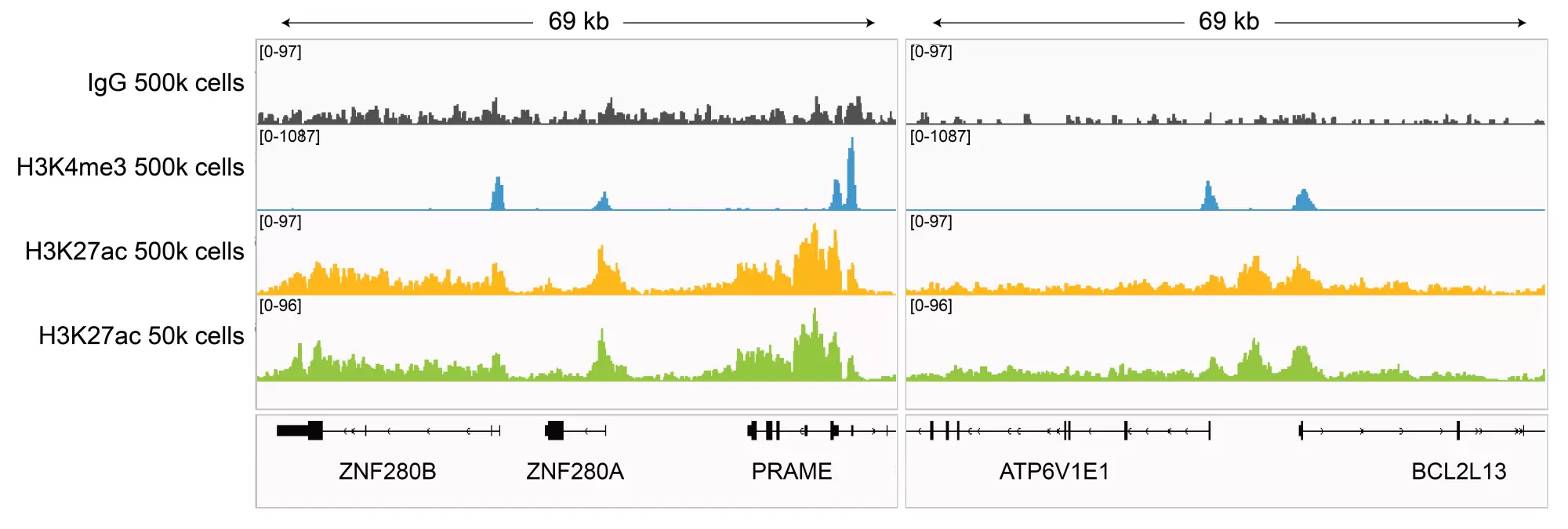
Figure 5: H3K27ac CUT&RUN representative browser tracks. CUT&RUN was performed and gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). H3K27ac antibody tracks display peaks at promoters and enhancers, consistent with the biological function of this PTM. Similar results in peak structure and location were observed for both 500k and 50k cell inputs.
Background References
[1] Pei et al. Clinical Epigenetics (2020). PMID: 32664951
[2] Simithy et al. Nature Communications (2017). PMID: 29070843
- Catalog Number
13-0059-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

