H3K36me3 Antibody, SNAP-Certified™ for CUT&RUN
CUTANA Compatible Antibodies meet the criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping.
Product Description
This H3K36me3 (histone H3 lysine 36 trimethyl) antibody meets EpiCypher’s lot-specific SNAP-Certified™ criteria for specificity and efficient target enrichment in both CUT&RUN and CUT&Tag applications. This requires <20% cross-reactivity to related histone PTMs determined using the SNAP-CUTANA™ K-MetStat Panel of spike-in controls (19-1002-EPC, Figure 1). High target efficiency is confirmed by consistent genomic enrichment at 500k and 50k starting cells (Figure 2). This antibody targets histone H3 trimethylated at lysine 36, which is enriched in promoters and gene bodies of active genes [1].
Validation Data
CUT&RUN was performed on 500k and 50k K562 cells with the SNAP-CUTANA™ K-MetStat Panel (19-1002-EPC) spiked-in prior to the addition of 0.5 µg of either IgG negative control (13-0042-EPC), H3K4me3 positive control (13-0041-EPC), or H3K36me3 antibodies. The experiment was performed using the CUTANA™ ChIC/CUT&RUN Kit v3.0 (14-1048-EPC). Library preparation was performed with 5 ng of CUT&RUN enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (14-1001-EPC/14-1002-EPC). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample sequencing depth was 16.8 million reads (IgG 500k cell input), 14.4 million reads (H3K4me3 500k cell input), 24.4 million reads (H3K36me3 500k cell input) and 16.4 million reads (H3K36me3 50k cell input). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and blacklist regions.
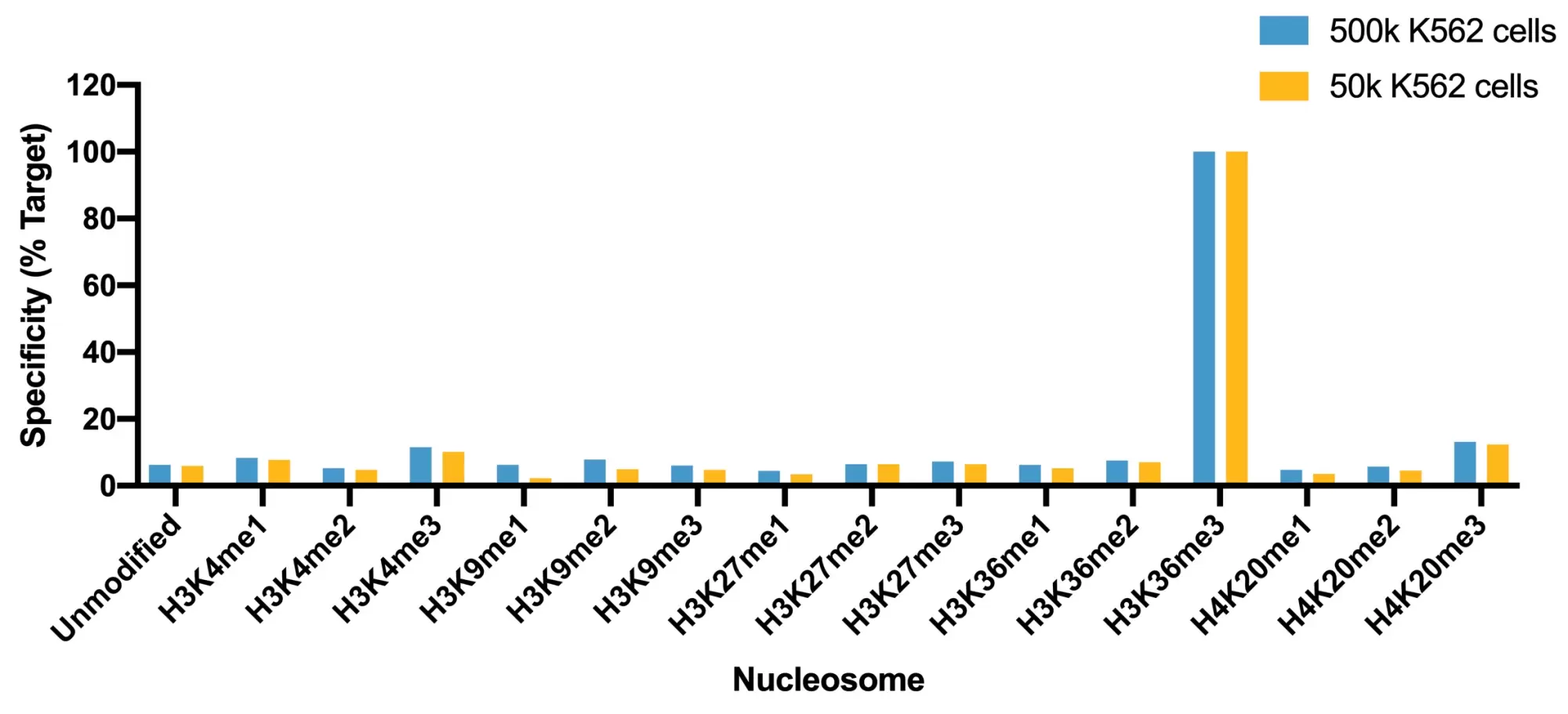
Figure 1: SNAP specificity analysis in CUT&RUN
CUT&RUN was performed as described above. CUT&RUN sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the K-MetStat panel (x-axis). Data are expressed as a percent relative to on-target recovery (H3K36me3 set to 100%).
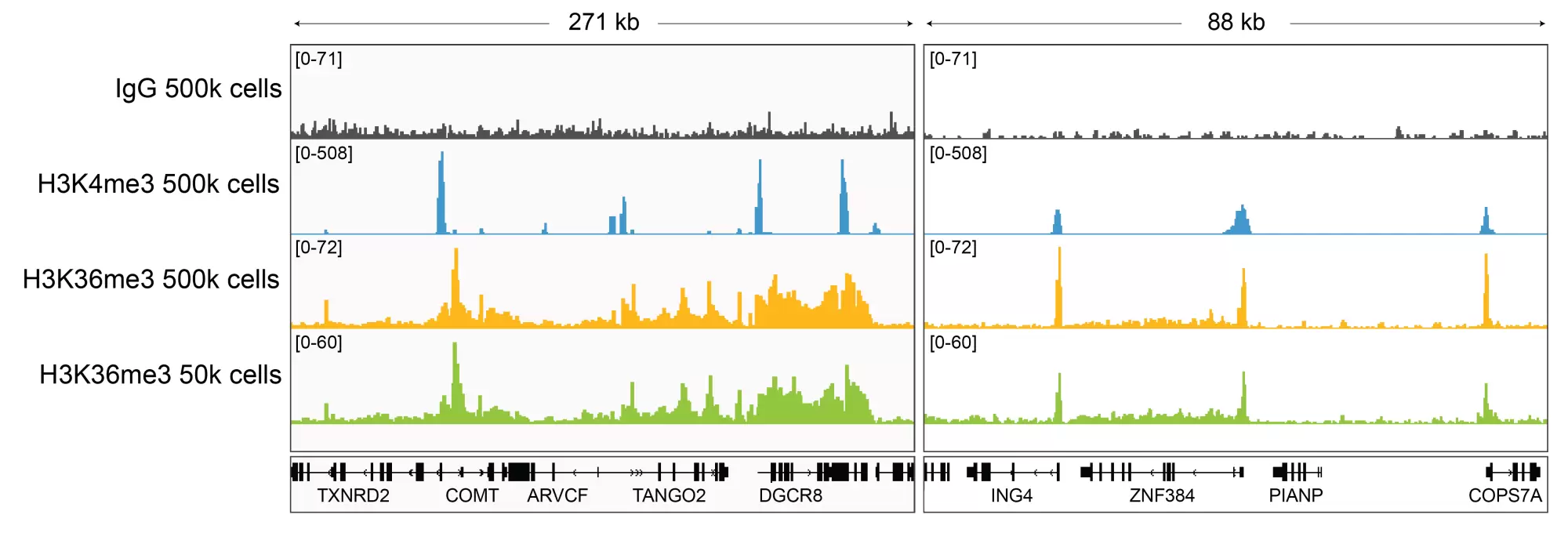
Figure 2: H3K36me3 CUT&RUN representative browser tracks
CUT&RUN was performed as described above. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). H3K36me3 antibody tracks display the characteristic enrichment known to be consistent with the function of this PTM [1]. Similar results in peak structure and location were observed for both 500k and 50k cell inputs.
Product Citations
- Joydeb Sinha, Jan F Nickels, Abby R Thurm, Connor H Ludwig, Bella N Archibald, Michaela M Hinks, Jun Wan, Dong Fang, Lacramioara Bintu (2024) The H3.3K36M oncohistone disrupts the establishment of epigenetic memory through loss of DNA methylation. Molecular cell
- Stephen L. McDaniel, Austin Hepperla, Jie Huang, Raghuvar Dronamraju, Alexander T. Adams, Vidyadhar G. Kulkarni, Ian J. Davis, Brian D. Strahl (2017) H3K36 Methylation Regulates Nutrient Stress Response in Saccharomyces cerevisiae by Enforcing Transcriptional Fidelity Cell reports
- Catalog Number
13-0058-EPC - Supplier
EpiCypher - Size
- Shipping
Blue Ice

