10X Visium Digital Spatial Gene Expression Service
Map the whole transcriptome within the tissue context with 10X Visium Digital Spatial Gene Expression
Product Description
No experimental setup, no delays, and no hassle!
10x Visium Spatial Gene Expression is a next generation molecular profiling solution for classifying tissue based on total mRNA. Mapping gene expression by spatially resolving the whole transcriptome allows you to determine the function of distinct cell populations, to explore how proximity affects biological response, to identify new biomarkers, and to profile distinct biological compartments.
- Flexible - suitable for both FFPE & Fresh-frozen tissues
- Comprehensive - analyze the whole transcriptome on entire tissue sections
- Adaptable - combine WTA with immunofluorescence protein detection
Unravel biological architectures in normal and diseased tissue and discover new biomarkers.
Please contact us to discuss your project and get a quote.
10x Visium Spatial Gene Expression Workflow
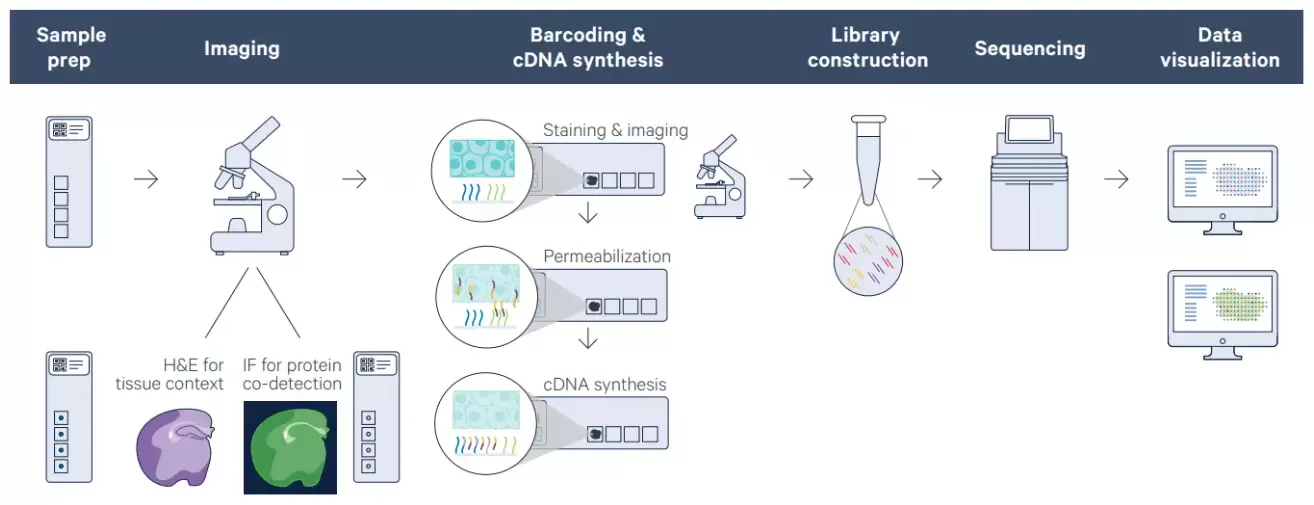
Spatial capture technology. mRNA molecules get a spatial barcode by the use of spatially barcoded mRNA – binding oligonucleotides.
Gain High-Resolution Characterization of Gene and Protein Expression
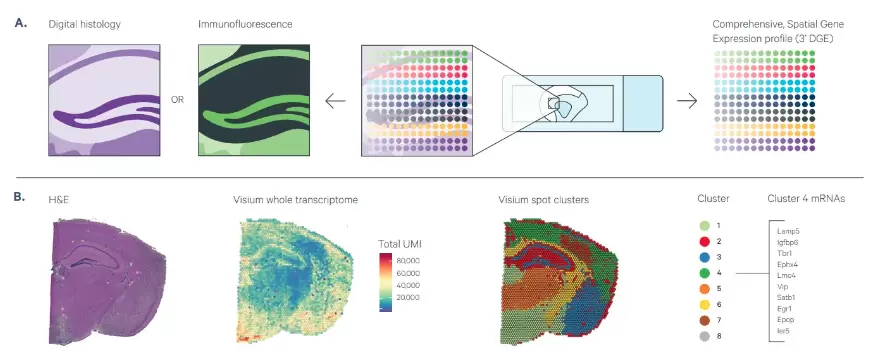
(A) Visium Spatial Gene Expression provides unbiased or targeted gene expression readout in intact sections from fresh-frozen tissue. The workflow is compatible with H&E to provide morphological context and IF staining to co-detect protein from the same tissue section.
(B) Shown on the left is an H&E image for a coronal mouse brain section, followed by an overlay of Visium data for total unique molecular identifiers (UMIs) for whole transcriptome analysis or spatially naïve spot clustering based on total differentially expressed genes. Listed on the far right are the most highly expressed genes in Cluster 4.
- Catalog Number
10XV-CS-BC - Supplier
BioChain Institute - Size
- Shipping
RT
- Price
- Please inquire

