CRISPR/Cas9 Lentiviral sgRNA Libraries
CRISPR/Cas9 Lentiviral sgRNA Libraries
CRISPR Knockout, CRISPRa, CRISPRi and CRISPR-seq
CRISPR Knockout Pooled Lentiviral sgRNA Libraries
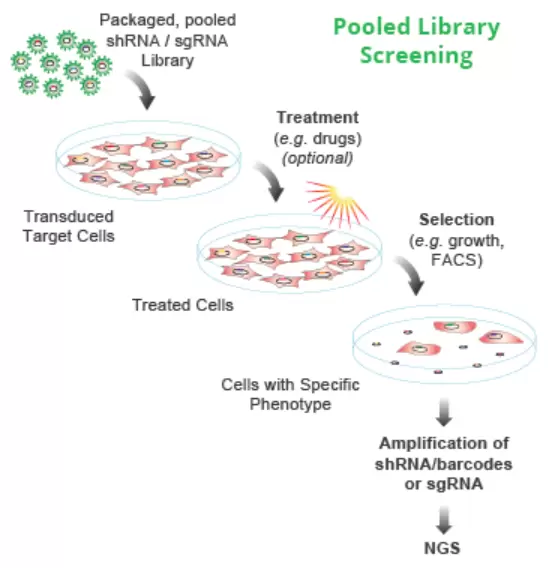
Quality pooled lentiviral CRISPR sgRNA libraries for unbiased phenotypic knockout screens in mammalian cell systems are provided. Pooled libraries of many thousands of defined single-guide RNA (sgRNA or gRNA) sequences enable disruption (or “knockout”) of thousands of genes throughout a cell population in a single experiment. Screening this cell population for a specific phenotype (e.g., viability) then, enables the identification of specific genes driving that phenotype, such as a drug response or disease phenotype.
- Effective guides incorporate Cellecta’s tracrRNA “HEAT” design and have been screened to minimize off-target knockout
- Both plasmid and pre-packaged lentivirus forms of the libraries are available
- Sufficient plasmid library is provided for immediate packaging (no re-amplification necessary)
Guide designs for Cellecta’s libraries incorporate state-of-the-art design criteria to ensure the most effective knockout possible. In addition, the region of the sgRNA interacting with the Cas9 nuclease has been optimized for higher efficiency knockout by incorporating the “HEAT” design. It has been shown that specific modifications to the Cas9-binding region of sgRNA significantly improve the effectiveness of gene knockout and that CRISPR libraries containing constructs with these modified sgRNA sequences generate stronger and more robust results than those with the standard sgRNA.
CRISPRa and CRISPRi Lentiviral sgRNA Libraries
Pre-made and custom pooled lentiviral libraries for both CRISPRa targeted gene activation and CRISPRi targeted gene repression screens are offered. These modified CRISPR platforms provide alternative genetic screening approaches that enable idenfication of genes whose modulation - either repression or activation - initiates a response.
- Off-the-shelf, genome-wide, lentiviral CRISPRa and CRISPRi libraries for human and mouse are available
- Libraries which comprise two sgRNAs targeting each gene for stronger activation and repression (dual-sgRNA constructs library) are only commercially available from Cellecta
- Both plasmid (200 ug) and pre-packaged lentiviral particle (2x 10^8 TU & 1x 10^9 TU) formats are available
- CRISPRa Dual-sgRNA libraries can be obtained with the following vectors: UbiC-RFP-Puro, UbiC-Puro or UbiC-RFP-Bleo
Engineered versions of deactivated Cas9 (dCas9) have proven to be effective modulators of gene expression when complexed with repressor (e.g., dCas9-KRAB) or activator (e.g., dCas9-VPH) proteins and targeted to the upstream promoter regions of gene targets with sgRNA. This technology forms the basis of the “CRISPR interference” (CRISPRi) and CRISPR activation (CRISPRa) systems.
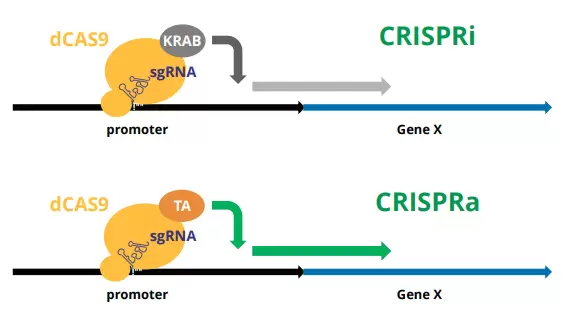
Cellecta’s CRISPRa and CRISPRi human genome-wide sgRNA libraries have been designed to target almost 19,000 human protein-coding genes using 5 sgRNAs designed against the promoter region based on Horlbeck, et al. (eLife 2016;5:e19760 DOI: 10.7554). The dual-guide CRISPRa and CRISPRi libraries use the same 5 effective sgRNA in dual combination on each of 5 constructs (sgRNA 1 & 2, sgRNA 1 & 3, sgRNA 2 & 3, etc.) Libraries are available in 200 ug plasmid and pre-packaged 2 x 10^8 TU and 1 x 10^9 TU lentiviral particles.
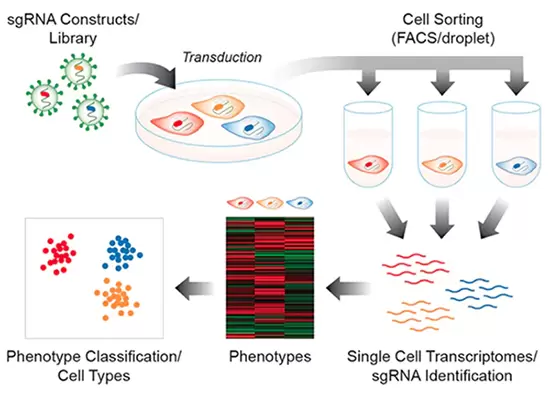
CRISPR-Seq Barcoded sgRNA Libraries (Perturb-Seq, CROP-Seq)
CloneTracker XP-CRISPR Libraries developed by Cellecta are pooled lentiviral libraries with a barcoded sgRNA expression cassette that is expressed and can be detected using RNA sequencing.
- Run CRISPR screens with single-cell RNA-sequencing platforms (aka Perturb-Seq or CRISP‑Seq)
- Identify gene activation changes in response to knockouts on a cell-to-cell basis
- Mouse and Human CloneTracker XP-CRISPR Libraries to anti-cancer targets, ready-to-use, and custom libraries for any set of genes
Pooled CRISPR screening with lentiviral sgRNA libraries has proven to be a very effective approach to identify genes functionally required to generate particular phenotypes, and RNA-Seq is an effective method to find the underlying changes in gene expression producing those phenotypes. With the advent of droplet microfluidic platforms that enable single-cell molecular analysis on a large scale, these two technologies may be merged to assess and characterize the distinct expression profiles produced by a genetic disruption in a pooled CRISPR knockout screen. With this sort of pooled screening approach using CRISPR barcode libraries, isolation and characterization of phenotype-specific cells are unnecessary.