
Methyl-seq Bisulfite Sequencing Services
Methyl-seq Services
Get rapidly and reliably to whole genome, reduced representation or targeted bisulfite DNA sequencing data
Genome-wide, base resolution DNA methylation analysis is now made easy using the comprehensive Methyl-Seq bisulfite sequencing services of our partner EpigenTek.
- Complete conversion (>99.9%) and maximum recovery of bisulfite-converted DNA
- Minimal selection bias, allowing for a low amount of input material high yield of the constructed DNA library, and low error rates
- Streamlined process for rapid turnaround time (4-9 weeks)
- Multi-stage quality control check including Bioanalyzer library QC and KAPA qPCR quantification
- Q30 > 75% sequencing score guaranteed
- End-to-end service with bioinformatics included at no additional charge
- Delivery of high-quality, publishable data
- Receive full deliverable package including Illumina FASTQ raw sequencing files, FastQC quality control insight, Bismark read mapping, methylation calling data, differential methylation analysis, and methylation region annotation
- Post-completion technical support to assist with data analysis
The next-generation sequencing platforms can deliver a great amount of useful DNA methylation information with publication-ready data parsed by our expert bioinformatics scientists. The study of methylation at single base resolution of individual cytosines in DNA is facilitated by bisulfite treatment of DNA followed by PCR amplification, cloning, and sequencing of individual amplimers.
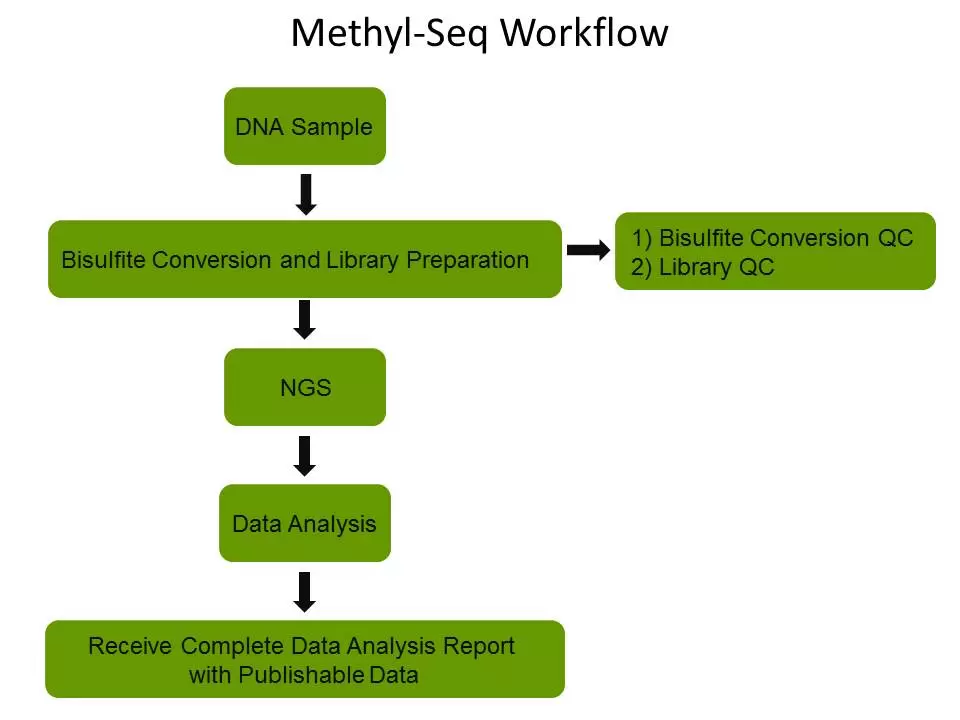
EpiGentek’s Methyl-Seq platform uses Targeted Bisulfite Sequencing, Reduced Representation Bisulfite Sequencing (RRBS) or Whole Genome Bisulfite Sequencing (WGBS) to yield reliable information on the methylation states of individual cytosines by effectively and efficiently preparing converted DNA for use in next generation sequencing techniques.